A structurally minimized yet fully active insulin based on cone-snail venom insulin principles
- PMID: 32483339
- PMCID: PMC7374640
- DOI: 10.1038/s41594-020-0430-8
A structurally minimized yet fully active insulin based on cone-snail venom insulin principles
Erratum in
-
Author Correction: A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.Nat Struct Mol Biol. 2020 Jul;27(7):683. doi: 10.1038/s41594-020-0460-2. Nat Struct Mol Biol. 2020. PMID: 32533129
Abstract
Human insulin and its current therapeutic analogs all show propensity, albeit varyingly, to self-associate into dimers and hexamers, which delays their onset of action and makes blood glucose management difficult for people with diabetes. Recently, we described a monomeric, insulin-like peptide in cone-snail venom with moderate human insulin-like bioactivity. Here, with insights from structural biology studies, we report the development of mini-Ins-a human des-octapeptide insulin analog-as a structurally minimal, full-potency insulin. Mini-Ins is monomeric and, despite the lack of the canonical B-chain C-terminal octapeptide, has similar receptor binding affinity to human insulin. Four mutations compensate for the lack of contacts normally made by the octapeptide. Mini-Ins also has similar in vitro insulin signaling and in vivo bioactivities to human insulin. The full bioactivity of mini-Ins demonstrates the dispensability of the PheB24-PheB25-TyrB26 aromatic triplet and opens a new direction for therapeutic insulin development.
Figures






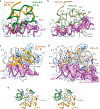
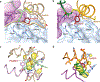




References
-
- Menting JG et al. A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin. Nat. Struct. Mol. Biol 23, 916–920 (2016). - PubMed
-
- Carpenter FH Relationship of structure to biological activity of insulin as revealed by degradative studies. Am. J. Med 40, 750–758 (1966). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

