C9orf72 suppresses systemic and neural inflammation induced by gut bacteria
- PMID: 32483373
- PMCID: PMC7416879
- DOI: 10.1038/s41586-020-2288-7
C9orf72 suppresses systemic and neural inflammation induced by gut bacteria
Abstract
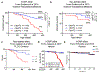
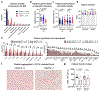
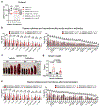
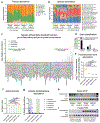
A hexanucleotide-repeat expansion in C9ORF72 is the most common genetic variant that contributes to amyotrophic lateral sclerosis and frontotemporal dementia1,2. The C9ORF72 mutation acts through gain- and loss-of-function mechanisms to induce pathways that are implicated in neural degeneration3-9. The expansion is transcribed into a long repetitive RNA, which negatively sequesters RNA-binding proteins5 before its non-canonical translation into neural-toxic dipeptide proteins3,4. The failure of RNA polymerase to read through the mutation also reduces the abundance of the endogenous C9ORF72 gene product, which functions in endolysosomal pathways and suppresses systemic and neural inflammation6-9. Notably, the effects of the repeat expansion act with incomplete penetrance in families with a high prevalence of amyotrophic lateral sclerosis or frontotemporal dementia, indicating that either genetic or environmental factors modify the risk of disease for each individual. Identifying disease modifiers is of considerable translational interest, as it could suggest strategies to diminish the risk of developing amyotrophic lateral sclerosis or frontotemporal dementia, or to slow progression. Here we report that an environment with reduced abundance of immune-stimulating bacteria10,11 protects C9orf72-mutant mice from premature mortality and significantly ameliorates their underlying systemic inflammation and autoimmunity. Consistent with C9orf72 functioning to prevent microbiota from inducing a pathological inflammatory response, we found that reducing the microbial burden in mutant mice with broad spectrum antibiotics-as well as transplanting gut microflora from a protective environment-attenuated inflammatory phenotypes, even after their onset. Our studies provide further evidence that the microbial composition of our gut has an important role in brain health and can interact in surprising ways with well-known genetic risk factors for disorders of the nervous system.
Conflict of interest statement
Conflicts of interest
KE is a co-founder of Q-State Biosciences, Quralis and Endear Therapies.
Figures








Comment in
-
Gut microbes tune inflammation and lifespan in a mouse model of amyotrophic lateral sclerosis.Nature. 2020 Jun;582(7810):34-35. doi: 10.1038/d41586-020-01335-3. Nature. 2020. PMID: 32405038 No abstract available.
-
A Gut Feeling about C9ORF72.Trends Immunol. 2020 Sep;41(9):755-757. doi: 10.1016/j.it.2020.07.012. Epub 2020 Aug 13. Trends Immunol. 2020. PMID: 32800706
References
-
- Mori K et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science 339, 1335–1338 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

