STIM1 is a metabolic checkpoint regulating the invasion and metastasis of hepatocellular carcinoma
- PMID: 32483465
- PMCID: PMC7255033
- DOI: 10.7150/thno.44025
STIM1 is a metabolic checkpoint regulating the invasion and metastasis of hepatocellular carcinoma
Erratum in
-
Erratum: STIM1 is a metabolic checkpoint regulating the invasion and metastasis of hepatocellular carcinoma: Erratum.Theranostics. 2025 Jun 15;15(14):7178-7179. doi: 10.7150/thno.119395. eCollection 2025. Theranostics. 2025. PMID: 40585985 Free PMC article.
Abstract
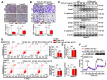
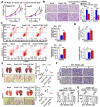
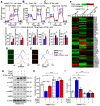
Background: Cancer cells undergoing invasion and metastasis possess a phenotype with attenuated glycolysis, but enhanced fatty acid oxidation (FAO). Calcium (Ca2+)-mediated signaling pathways are implicated in tumor metastasis and metabolism regulation. Stromal-interaction molecule 1 (STIM1) triggered store-operated Ca2+ entry (SOCE) is the major route of Ca2+ influx for non-excitable cells including hepatocellular carcinoma (HCC) cells. However, whether and how STIM1 regulates the invasion and metastasis of HCC via metabolic reprogramming is unclear. Methods: The expressions of STIM1 and Snail1 in the HCC tissues and cells were measured by immunohistochemistry, Western-blotting and quantitative PCR. STIM1 knockout-HCC cells were generated by CRISPR-Cas9, and gene-overexpression was mediated via lentivirus transfection. Besides, the invasive and metastatic activities of HCC cells were assessed by transwell assay, anoikis rate in vitro and lung metastasis in vivo. Seahorse energy analysis and micro-array were used to evaluate the glucose and lipid metabolism. Results: STIM1 was down-regulated in metastatic HCC cells rather than in proliferating HCC cells, and low STIM1 levels were associated with poor outcome of HCC patients. During tumor growth, STIM1 stabilized Snail1 protein by activating the CaMKII/AKT/GSK-3β pathway. Subsequently, the upregulated Snail1 suppressed STIM1/SOCE during metastasis. STIM1 restoration significantly diminished anoikis-resistance and metastasis induced by Snail1. Mechanistically, the downregulated STIM1 shifted the anabolic/catabolic balance, i.e., from aerobic glycolysis towards AMPK-activated fatty acid oxidation (FAO), which contributed to Snail1-driven metastasis and anoikis-resistance. Conclusions: Our data provide the molecular basis that STIM1 orchestrates invasion and metastasis via reprogramming HCC metabolism.
Keywords: SOCE; STIM1; Snail1; invasion and metastasis; metabolic reprogramming.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Cha YH, Yook JI, Kim HS, Kim NH. Catabolic metabolism during cancer EMT. Arch Pharm Res. 2015;38:313–20. - PubMed
-
- Herst PM, Grasso C, Berridge MV. Metabolic reprogramming of mitochondrial respiration in metastatic cancer. Cancer Metastasis Rev. 2018;37:643–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

