Multiple mutations at exon 2 of RHOA detected in plasma from patients with peripheral T-cell lymphoma
- PMID: 32484856
- PMCID: PMC7284097
- DOI: 10.1182/bloodadvances.2019001075
Multiple mutations at exon 2 of RHOA detected in plasma from patients with peripheral T-cell lymphoma
Abstract
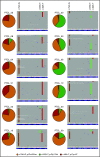
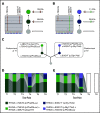
The mutational landscape of peripheral T-cell lymphoma (PTCL) is being revealed through sequencing of lymph node samples, but there has been little work on the mutational load that is present in cell-free DNA (cfDNA) from plasma. We report targeted sequencing of cfDNA from PTCL patients to demonstrate c.50G>T (p.Gly17Val) in RHOA as previously described in angioimmunoblastic T-cell lymphoma (AITL) and a group of PTCL not otherwise specified (NOS) but also detect novel mutations at c.73A>G (p.Phe25Leu) and c.48A>T (p.Cys16*) of exon 2, which were confirmed by Sanger sequencing. In a group of AITL and PTCL-NOS analyzed by droplet digital polymerase chain reaction, 63% (12/19) showed c.50G>T (p.Gly17Val), 53% (10/19) c.73A>G (p.Phe25Leu), and 37% (7/19) c.48A>T (pCys16*). Sequencing of lymph node tissue in 3 out of 9 cases confirmed the presence of c.73A>G (p.Phe25Leu). Inspection of individual sequencing reads from individual patients showed that a single RHOA allele could contain >1 mutation, suggesting haplotypes of mutations at RHOA. Serial sampling showed changes to RHOA mutational frequency with treatment and the apparent occurrence of clones bearing specific haplotypes associated with relapse. Therefore, sequencing of RHOA from cfDNA has revealed new mutations and haplotypes. The clinical significance of these findings will need to be explored in clinical trials, but liquid biopsy might have potential for guiding treatment decisions in PTCL.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




References
-
- Foss FM, Zinzani PL, Vose JM, Gascoyne RD, Rosen ST, Tobinai K. Peripheral T-cell lymphoma. Blood. 2011;117(25):6756-6767. - PubMed
-
- Vose J, Armitage J, Weisenburger D; International T-Cell Lymphoma Project . International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26(25):4124-4130. - PubMed
-
- de Leval L, Rickman DS, Thielen C, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109(11):4952-4963. - PubMed
-
- Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366(1):95-96. - PubMed

