Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic Pathogen Klebsiella oxytoca
- PMID: 32485104
- PMCID: PMC7354218
- DOI: 10.1021/acsinfecdis.0c00326
Biosynthesis, Mechanism of Action, and Inhibition of the Enterotoxin Tilimycin Produced by the Opportunistic Pathogen Klebsiella oxytoca
Abstract
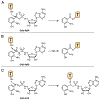
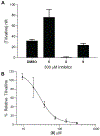
Tilimycin is an enterotoxin produced by the opportunistic pathogen Klebsiella oxytoca that causes antibiotic-associated hemorrhagic colitis (AAHC). This pyrrolobenzodiazepine (PBD) natural product is synthesized by a bimodular nonribosomal peptide synthetase (NRPS) pathway composed of three proteins: NpsA, ThdA, and NpsB. We describe the functional and structural characterization of the fully reconstituted NRPS system and report the steady-state kinetic analysis of all natural substrates and cofactors as well as the structural characterization of both NpsA and ThdA. The mechanism of action of tilimycin was confirmed using DNA adductomics techniques through the detection of putative N-2 guanine alkylation after tilimycin exposure to eukaryotic cells, providing the first structural characterization of a PBD-DNA adduct formed in cells. Finally, we report the rational design of small-molecule inhibitors that block tilimycin biosynthesis in whole cell K. oxytoca (IC50 = 29 ± 4 μM) through the inhibition of NpsA (KD = 29 ± 4 nM).
Keywords: Klebsiella oxytoca; adenylation; microbiome; nonribosomal peptide synthetase; pyrrolobenzodiazepine; tilimycin.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
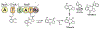
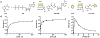
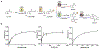
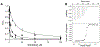







References
-
- Savage DC (1977) Microbial Ecology of the Gastrointestinal Tract. Annu. Rev. Microbiol 31, 107–133. - PubMed
-
- Nougayrede J-P (2006) Escherichia coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science 313, 848–851. - PubMed
-
- Peery AF, Dellon ES, Lund J, Crockett SD, McGowan CE, Bulsiewicz WJ, Gangarosa LM, Thiny MT, Stizenberg K, Morgan DR, Ringel Y, Kim HP, DiBonaventura MD, Carroll CF, Allen JK, Cook SF, Sandler RS, Kappelman MD, and Shaheen NJ (2012) Burden of Gastrointestinal Disease in the United States: 2012 Update. Gastroenterology 143, 1179–1187.e3. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

