Current View on EpCAM Structural Biology
- PMID: 32486423
- PMCID: PMC7349879
- DOI: 10.3390/cells9061361
Current View on EpCAM Structural Biology
Abstract
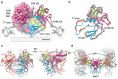
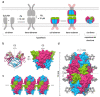
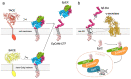
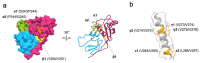
EpCAM, a carcinoma cell-surface marker protein and a therapeutic target, has been primarily addressed as a cell adhesion molecule. With regard to recent discoveries of its role in signaling with implications in cell proliferation and differentiation, and findings contradicting a direct role in mediating adhesion contacts, we provide a comprehensive and updated overview on the available structural data on EpCAM and interpret it in the light of recent reports on its function. First, we describe the structure of extracellular part of EpCAM, both as a subunit and part of a cis-dimer which, according to several experimental observations, represents a biologically relevant oligomeric state. Next, we provide a thorough evaluation of reports on EpCAM as a homophilic cell adhesion molecule with a structure-based explanation why direct EpCAM participation in cell-cell contacts is highly unlikely. Finally, we review the signaling aspect of EpCAM with focus on accessibility of signaling-associated cleavage sites.
Keywords: Keywords: EpCAM; adhesion; dimer; disease; regulated intramembrane proteolysis; signaling; structure; transmembrane protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Murakami N., Mori T., Nakamura S., Yoshimoto S., Honma Y., Ueno T., Kobayashi K., Kashihara T., Takahashi K., Inaba K., et al. Prognostic value of the expression of epithelial cell adhesion molecules in head and neck squamous cell carcinoma treated by definitive radiotherapy. J. Radiat. Res. 2019;60:803–811. doi: 10.1093/jrr/rrz053. - DOI - PMC - PubMed
-
- Tsaktanis T., Kremling H., Pavšič M., Von Stackelberg R., Mack B., Fukumori A., Steiner H., Vielmuth F., Spindler V., Huang Z., et al. Cleavage and Cell Adhesion Properties of Human Epithelial Cell Adhesion Molecule (HEPCAM)*. J. Boil. Chem. 2015;290:24574–24591. doi: 10.1074/jbc.M115.662700. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

