Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows
- PMID: 32487174
- PMCID: PMC7265231
- DOI: 10.1186/s13059-020-02048-6
Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows
Abstract
Background: Single-cell RNA sequencing has been widely adopted to estimate the cellular composition of heterogeneous tissues and obtain transcriptional profiles of individual cells. Multiple approaches for optimal sample dissociation and storage of single cells have been proposed as have single-nuclei profiling methods. What has been lacking is a systematic comparison of their relative biases and benefits.
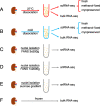
Results: Here, we compare gene expression and cellular composition of single-cell suspensions prepared from adult mouse kidney using two tissue dissociation protocols. For each sample, we also compare fresh cells to cryopreserved and methanol-fixed cells. Lastly, we compare this single-cell data to that generated using three single-nucleus RNA sequencing workflows. Our data confirms prior reports that digestion on ice avoids the stress response observed with 37 °C dissociation. It also reveals cell types more abundant either in the cold or warm dissociations that may represent populations that require gentler or harsher conditions to be released intact. For cell storage, cryopreservation of dissociated cells results in a major loss of epithelial cell types; in contrast, methanol fixation maintains the cellular composition but suffers from ambient RNA leakage. Finally, cell type composition differences are observed between single-cell and single-nucleus RNA sequencing libraries. In particular, we note an underrepresentation of T, B, and NK lymphocytes in the single-nucleus libraries.
Conclusions: Systematic comparison of recovered cell types and their transcriptional profiles across the workflows has highlighted protocol-specific biases and thus enables researchers starting single-cell experiments to make an informed choice.
Keywords: 10x Genomics; RNA-seq; Single-cell transcriptomics; scRNA-seq; snRNA-seq.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Combes AN, Phipson B, Lawlor KT, Dorison A, Patrick R, Zappia L, et al. Single cell analysis of the developing mouse kidney provides deeper insight into marker gene expression and ligand-receptor crosstalk. Development. 2019;146:dev178673. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

