Using soil bacterial communities to predict physico-chemical variables and soil quality
- PMID: 32487269
- PMCID: PMC7268603
- DOI: 10.1186/s40168-020-00858-1
Using soil bacterial communities to predict physico-chemical variables and soil quality
Abstract
Background: Soil ecosystems consist of complex interactions between biological communities and physico-chemical variables, all of which contribute to the overall quality of soils. Despite this, changes in bacterial communities are ignored by most soil monitoring programs, which are crucial to ensure the sustainability of land management practices. We applied 16S rRNA gene sequencing to determine the bacterial community composition of over 3000 soil samples from 606 sites in New Zealand. Sites were classified as indigenous forests, exotic forest plantations, horticulture, or pastoral grasslands; soil physico-chemical variables related to soil quality were also collected. The composition of soil bacterial communities was then used to predict the land use and soil physico-chemical variables of each site.
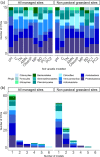
Results: Soil bacterial community composition was strongly linked to land use, to the extent where it could correctly determine the type of land use with 85% accuracy. Despite the inherent variation introduced by sampling across ~ 1300 km distance gradient, the bacterial communities could also be used to differentiate sites grouped by key physico-chemical properties with up to 83% accuracy. Further, individual soil variables such as soil pH, nutrient concentrations and bulk density could be predicted; the correlations between predicted and true values ranged from weak (R2 value = 0.35) to strong (R2 value = 0.79). These predictions were accurate enough to allow bacterial communities to assign the correct soil quality scores with 50-95% accuracy.
Conclusions: The inclusion of biological information when monitoring soil quality is crucial if we wish to gain a better, more accurate understanding of how land management impacts the soil ecosystem. We have shown that soil bacterial communities can provide biologically relevant insights on the impacts of land use on soil ecosystems. Furthermore, their ability to indicate changes in individual soil parameters shows that analysing bacterial DNA data can be used to screen soil quality. Video Abstract.
Keywords: Bacterial communities; Bacterial indicators; Biomonitoring; Environmental monitoring; Random forest analysis; Soil health; Soil microbiology.
Conflict of interest statement
The authors declare that they have no competing interests
Figures





References
-
- Doran JW, Zeiss MR (2000) Soil health and sustainability: managing the biotic component of soil quality. Appl Soil Ecol 15:3–11. 10.1016/S0929-1393(00)00067-6.
-
- Oliver MA, Gregory PJ. Soil, food security and human health: a review. Eur J Soil Sci. 2015;66:257–276. doi: 10.1111/ejss.12216. - DOI
-
- Wolters V (2001) Biodiversity of soil animals and its function. Eur J Soil Biol 37:221–227. 10.1016/S1164-5563(01)01088-3.
-
- Ranjard L, Poly F, Nazaret S (2000) Monitoring complex bacterial communities using culture-independent molecular techniques: application to soil environment. Res Microbiol 151:167–177. 10.1016/S0923-2508(00)00136-4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

