Supernumerary B chromosomes of Aegilops speltoides undergo precise elimination in roots early in embryo development
- PMID: 32488019
- PMCID: PMC7265534
- DOI: 10.1038/s41467-020-16594-x
Supernumerary B chromosomes of Aegilops speltoides undergo precise elimination in roots early in embryo development
Abstract
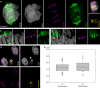
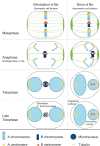
Not necessarily all cells of an organism contain the same genome. Some eukaryotes exhibit dramatic differences between cells of different organs, resulting from programmed elimination of chromosomes or their fragments. Here, we present a detailed analysis of programmed B chromosome elimination in plants. Using goatgrass Aegilops speltoides as a model, we demonstrate that the elimination of B chromosomes is a strictly controlled and highly efficient root-specific process. At the onset of embryo differentiation B chromosomes undergo elimination in proto-root cells. Independent of centromere activity, B chromosomes demonstrate nondisjunction of chromatids and lagging in anaphase, leading to micronucleation. Chromatin structure and DNA replication differ between micronuclei and primary nuclei and degradation of micronucleated DNA is the final step of B chromosome elimination. This process might allow root tissues to survive the detrimental expression, or overexpression of B chromosome-located root-specific genes with paralogs located on standard chromosomes.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Smith JJ. Programmed DNA elimination: keeping germline genes in their place. Curr. Biol. 2018;28:R601–R603. - PubMed
-
- Marcussen T, et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345:1250092. - PubMed
-
- Mendelson D, Zohary D. Behavior and transmission of supernumerary chromosomes in Aegilops speltoides. Heredity. 1972;29:329–339.
-
- Jones RN, Rees H. B chromosomes. New York: Academic Press; 1982.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

