Critical Roles of Translation Initiation and RNA Uridylation in Endogenous Retroviral Expression and Neural Differentiation in Pluripotent Stem Cells
- PMID: 32492424
- PMCID: PMC8195978
- DOI: 10.1016/j.celrep.2020.107715
Critical Roles of Translation Initiation and RNA Uridylation in Endogenous Retroviral Expression and Neural Differentiation in Pluripotent Stem Cells
Abstract
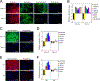
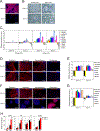
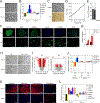
Previous studies have suggested that the loss of the translation initiation factor eIF4G1 homolog NAT1 induces excessive self-renewability of naive pluripotent stem cells (PSCs); yet the role of NAT1 in the self-renewal and differentiation of primed PSCs is still unclear. Here, we generate a conditional knockout of NAT1 in primed PSCs and use the cells for the functional analyses of NAT1. Our results show that NAT1 is required for the self-renewal and neural differentiation of primed PSCs. In contrast, NAT1 deficiency in naive pluripotency attenuates the differentiation to all cell types. We also find that NAT1 is involved in efficient protein expression of an RNA uridyltransferase, TUT7. TUT7 is involved in the neural differentiation of primed PSCs via the regulation of human endogenous retrovirus accumulation. These data demonstrate the essential roles of NAT1 and TUT7 in the precise transition of stem cell fate.
Keywords: endogenous retrovirus; neural differentiation; pluripotency; translation.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests K.T. is on the scientific advisory board of I Peace, without salary. B.R.C. is a scientific advisor to Tenaya Therapeutics. S.Y. is a scientific advisor (without salary) to iPS Academia Japan.
Figures




References
-
- Aronesty E (2013). Comparison of Sequencing Utility Programs. The Open Bioinformatics Journal 7, 1–8.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

