Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples
- PMID: 32492531
- PMCID: PMC7833505
- DOI: 10.1016/j.ijid.2020.05.099
Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples
Abstract
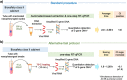
Objectives: The gold-standard COVID-19 diagnosis relies on detecting SARS-CoV-2 using RNA purification and one-step retrotranscription and quantitative PCR (RT-qPCR). Based on the urgent need for high-throughput screening, we tested the performance of three alternative, simple and affordable protocols to rapidly detect SARS-CoV-2, bypassing the long and tedious RNA extraction step and reducing the time to viral detection.
Methods: We evaluated three methods based on direct nasopharyngeal swab viral transmission medium (VTM) heating before the RT-qPCR: a) direct without additives; b) in a formamide-EDTA (FAE) buffer, c) in a RNAsnapTM buffer.
Results: Although with a delay in cycle threshold compared to the gold-standard, we found consistent results in nasopharyngeal swab samples that were subject to a direct 70°C incubation for 10 min.
Conclusions: Our findings provide valuable options to overcome any supply chain issue and help to increase the throughput of diagnostic tests, thereby complementing standard diagnosis.
Keywords: COVID-19; RNA extraction; SARS-CoV-2; diagnosis; fast protocols; sample treatment.
Copyright © 2020 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Sensitivity of different RT-qPCR solutions for SARS-CoV-2 detection.Int J Infect Dis. 2020 Oct;99:190-192. doi: 10.1016/j.ijid.2020.07.058. Epub 2020 Aug 1. Int J Infect Dis. 2020. PMID: 32745627 Free PMC article.
-
Laboratory management for SARS-CoV-2 detection: a user-friendly combination of the heat treatment approach and rt-Real-time PCR testing.Emerg Microbes Infect. 2020 Dec;9(1):1393-1396. doi: 10.1080/22221751.2020.1775500. Emerg Microbes Infect. 2020. PMID: 32552549 Free PMC article.
-
Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step.PLoS Biol. 2020 Oct 2;18(10):e3000896. doi: 10.1371/journal.pbio.3000896. eCollection 2020 Oct. PLoS Biol. 2020. PMID: 33006983 Free PMC article.
-
Point-of-Care Diagnostics of COVID-19: From Current Work to Future Perspectives.Sensors (Basel). 2020 Jul 31;20(15):4289. doi: 10.3390/s20154289. Sensors (Basel). 2020. PMID: 32752043 Free PMC article. Review.
-
SARS-CoV-2 detection in different respiratory sites: A systematic review and meta-analysis.EBioMedicine. 2020 Sep;59:102903. doi: 10.1016/j.ebiom.2020.102903. Epub 2020 Jul 24. EBioMedicine. 2020. PMID: 32718896 Free PMC article.
Cited by
-
Performance Evaluation of Different RT-PCR Kits for the Direct Detection of SARS-CoV-2 in Preheated Specimens.J Lab Physicians. 2023 Jan 30;15(3):383-391. doi: 10.1055/s-0043-1760752. eCollection 2023 Sep. J Lab Physicians. 2023. PMID: 37564223 Free PMC article.
-
Wide Application of Minimally Processed Saliva on Multiple RT-qPCR Kits for SARS-CoV-2 Detection in Indonesia.Front Cell Infect Microbiol. 2021 Aug 18;11:691538. doi: 10.3389/fcimb.2021.691538. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34485174 Free PMC article.
-
Extraction-free protocol combining proteinase K and heat inactivation for detection of SARS-CoV-2 by RT-qPCR.PLoS One. 2021 Feb 26;16(2):e0247792. doi: 10.1371/journal.pone.0247792. eCollection 2021. PLoS One. 2021. PMID: 33635936 Free PMC article.
-
Saliva: What Dental Practitioners Should Know about the Role of This Biofluid in the Transmission and Diagnostic of SARS-CoV-2.Medicina (Kaunas). 2021 Apr 6;57(4):349. doi: 10.3390/medicina57040349. Medicina (Kaunas). 2021. PMID: 33917276 Free PMC article. Review.
-
Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials.J Clin Virol. 2020 Sep;130:104579. doi: 10.1016/j.jcv.2020.104579. Epub 2020 Aug 5. J Clin Virol. 2020. PMID: 32795959 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

