Development and Application of a Core Genome Multilocus Sequence Typing Scheme for the Health Care-Associated Pathogen Pseudomonas aeruginosa
- PMID: 32493782
- PMCID: PMC7448633
- DOI: 10.1128/JCM.00214-20
Development and Application of a Core Genome Multilocus Sequence Typing Scheme for the Health Care-Associated Pathogen Pseudomonas aeruginosa
Abstract
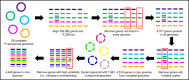
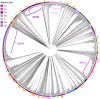
Pseudomonas aeruginosa is an opportunistic human pathogen that frequently causes health care-associated infections (HAIs). Due to its metabolic diversity and ability to form biofilms, this Gram-negative nonfermenting bacterium can persist in the health care environment, which can lead to prolonged HAI outbreaks. We describe the creation of a core genome multilocus sequence typing (cgMLST) scheme to provide a stable platform for the rapid comparison of P. aeruginosa isolates using whole-genome sequencing (WGS) data. We used a diverse set of 58 complete P. aeruginosa genomes to curate a set of 4,440 core genes found in each isolate, representing ∼64% of the average genome size. We then expanded the alleles for each gene using 1,991 contig-level genome sequences. The scheme was used to analyze genomes from four historical HAI outbreaks to compare the phylogenies generated using cgMLST to those of other means (traditional MLST, pulsed-field gel electrophoresis [PFGE], and single-nucleotide variant [SNV] analysis). The cgMLST scheme provides sufficient resolution for analyzing individual outbreaks, as well as the stability for comparisons across a variety of isolates encountered in surveillance studies, making it a valuable tool for the rapid analysis of P. aeruginosa genomes.
Keywords: HAI; Pseudomonas aeruginosa; antibiotic resistance; cgMLST.
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Figures


References
-
- 2019. AR threats report. (Accessed Nov. 22, 2019). https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-re....
-
- Burdette SD, Herchline TE. 2006. Antimicrobe.org: an online reference for the practicing infectious diseases specialist. Clin Infect Dis 43:765–769. doi:10.1086/507039. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

