Ebola Virus Inclusion Body Formation and RNA Synthesis Are Controlled by a Novel Domain of Nucleoprotein Interacting with VP35
- PMID: 32493824
- PMCID: PMC7394894
- DOI: 10.1128/JVI.02100-19
Ebola Virus Inclusion Body Formation and RNA Synthesis Are Controlled by a Novel Domain of Nucleoprotein Interacting with VP35
Abstract
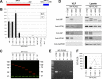
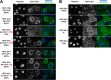
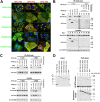
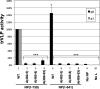
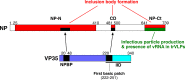
Ebola virus (EBOV) inclusion bodies (IBs) are cytoplasmic sites of nucleocapsid formation and RNA replication, housing key steps in the virus life cycle that warrant further investigation. During infection, IBs display dynamic properties regarding their size and location. The contents of IBs also must transition prior to further viral maturation, assembly, and release, implying additional steps in IB function. Interestingly, the expression of the viral nucleoprotein (NP) alone is sufficient for the generation of IBs, indicating that it plays an important role in IB formation during infection. In addition to NP, other components of the nucleocapsid localize to IBs, including VP35, VP24, VP30, and the RNA polymerase L. We previously defined and solved the crystal structure of the C-terminal domain of NP (NP-Ct), but its role in virus replication remained unclear. Here, we show that NP-Ct is necessary for IB formation when NP is expressed alone. Interestingly, we find that NP-Ct is also required for the production of infectious virus-like particles (VLPs), and that defective VLPs with NP-Ct deletions are significantly reduced in viral RNA content. Furthermore, coexpression of the nucleocapsid component VP35 overcomes deletion of NP-Ct in triggering IB formation, demonstrating a functional interaction between the two proteins. Of all the EBOV proteins, only VP35 is able to overcome the defect in IB formation caused by the deletion of NP-Ct. This effect is mediated by a novel protein-protein interaction between VP35 and NP that controls both regulation of IB formation and RNA replication itself and that is mediated by a newly identified functional domain of NP, the central domain.IMPORTANCE Inclusion bodies (IBs) are cytoplasmic sites of RNA synthesis for a variety of negative-sense RNA viruses, including Ebola virus. In addition to housing important steps in the viral life cycle, IBs protect new viral RNA from innate immune attack and contain specific host proteins whose function is under study. A key viral factor in Ebola virus IB formation is the nucleoprotein, NP, which also is important in RNA encapsidation and synthesis. In this study, we have identified two domains of NP that control inclusion body formation. One of these, the central domain (CD), interacts with viral protein VP35 to control both inclusion body formation and RNA synthesis. The other is the NP C-terminal domain (NP-Ct), whose function has not previously been reported. These findings contribute to a model in which NP and its interactions with VP35 link the establishment of IBs to the synthesis of viral RNA.
Keywords: RNA replication; ebola virus; inclusion body; nucleoprotein.
Copyright © 2020 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

