Depth-dependent mycoplankton glycoside hydrolase gene activity in the open ocean-evidence from the Tara Oceans eukaryote metatranscriptomes
- PMID: 32494052
- PMCID: PMC7608184
- DOI: 10.1038/s41396-020-0687-2
Depth-dependent mycoplankton glycoside hydrolase gene activity in the open ocean-evidence from the Tara Oceans eukaryote metatranscriptomes
Abstract
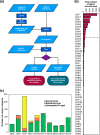
Mycoplankton are widespread components of marine ecosystems, yet the full extent of their functional role remains poorly known. Marine mycoplankton are likely functionally analogous to their terrestrial counterparts, including performing saprotrophy and degrading high-molecular weight organic substrates using carbohydrate-active enzymes (CAZymes). We investigated the prevalence of transcribed oceanic fungal CAZyme genes using the Marine Atlas of Tara Ocean Unigenes database. We revealed an abundance of unique transcribed fungal glycoside hydrolases in the open ocean, including a particularly high number that act upon cellulose in surface waters and the deep chlorophyll maximum (DCM). A variety of other glycoside hydrolases acting on a range of biogeochemically important polysaccharides including β-glucans and chitin were also found. This analysis demonstrates that mycoplankton are active saprotrophs in the open ocean and paves the way for future research into the depth-dependent roles of marine fungi in oceanic carbon cycling, including the biological carbon pump.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Cunliffe M, Hollingsworth A, Bain C, Sharma V, Taylor JD. Algal polysaccharide utilisation by saprotrophic planktonic marine fungi. Fungal Ecol. 2017;30:135–8. doi: 10.1016/j.funeco.2017.08.009. - DOI

