O-GlcNAc transferase promotes influenza A virus-induced cytokine storm by targeting interferon regulatory factor-5
- PMID: 32494619
- PMCID: PMC7159909
- DOI: 10.1126/sciadv.aaz7086
O-GlcNAc transferase promotes influenza A virus-induced cytokine storm by targeting interferon regulatory factor-5
Abstract
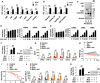
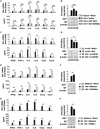
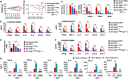
In this study, we demonstrated an essential function of the hexosamine biosynthesis pathway (HBP)-associated O-linked β-N-acetylglucosamine (O-GlcNAc) signaling in influenza A virus (IAV)-induced cytokine storm. O-GlcNAc transferase (OGT), a key enzyme for protein O-GlcNAcylation, mediated IAV-induced cytokine production. Upon investigating the mechanisms driving this event, we determined that IAV induced OGT to bind to interferon regulatory factor-5 (IRF5), leading to O-GlcNAcylation of IRF5 on serine-430. O-GlcNAcylation of IRF5 is required for K63-linked ubiquitination of IRF5 and subsequent cytokine production. Analysis of clinical samples revealed that IRF5 is O-GlcNAcylated, and higher levels of proinflammatory cytokines correlated with higher levels of blood glucose in IAV-infected patients. We identified a molecular mechanism by which HBP-mediated O-GlcNAcylation regulates IRF5 function during IAV infection, highlighting the importance of glucose metabolism in IAV-induced cytokine storm.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Yang Y. R., Song M., Lee H., Jeon Y., Choi E.-J., Jang H.-J., Moon H. Y., Byun H.-Y., Kim E.-K., Kim D. H., Lee M. N., Koh A., Ghim J., Choi J. H., Lee-Kwon W., Kim K. T., Ryu S. H., Suh P.-G., O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

