Cellular contractile forces are nonmechanosensitive
- PMID: 32494649
- PMCID: PMC7176410
- DOI: 10.1126/sciadv.aaz6997
Cellular contractile forces are nonmechanosensitive
Abstract
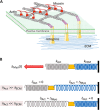
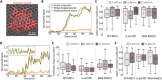
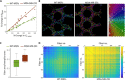
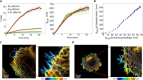
Cells' ability to apply contractile forces to their environment and to sense its mechanical properties (e.g., rigidity) are among their most fundamental features. Yet, the interrelations between contractility and mechanosensing, in particular, whether contractile force generation depends on mechanosensing, are not understood. We use theory and extensive experiments to study the time evolution of cellular contractile forces and show that they are generated by time-dependent actomyosin contractile displacements that are independent of the environment's rigidity. Consequently, contractile forces are nonmechanosensitive. We further show that the force-generating displacements are directly related to the evolution of the actomyosin network, most notably to the time-dependent concentration of F-actin. The emerging picture of force generation and mechanosensitivity offers a unified framework for understanding contractility.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures

References
-
- Prager-Khoutorsky M., Lichtenstein A., Krishnan R., Rajendran K., Mayo A., Kam Z., Geiger B., Bershadsky A. D., Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 13, 1457–1465 (2011). - PubMed
-
- Wang H.-B., Dembo M., Wang Y.-L., Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell Physiol. 279, C1345–C1350 (2000). - PubMed
-
- Chaudhuri O., Koshy S. T., Branco da Cunha C., Shin J. W., Verbeke C. S., Allison K. H., Mooney D. J., Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 13, 970–978 (2014). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

