Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis
- PMID: 32495982
- PMCID: PMC7419039
- DOI: 10.1111/cas.14511
Circular RNA LPAR3 sponges microRNA-198 to facilitate esophageal cancer migration, invasion, and metastasis
Abstract
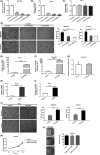
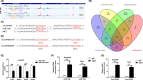
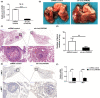
In this study, we explored expression and functions of circular RNA LPAR3 (circLPAR3) in esophageal squamous cell carcinoma (ESCC). The differential expression of circular RNAs (circRNAs) in 10 ESCC and corresponding paracarcinoma tissues was analyzed through circRNA microarray, then the candidate circRNAs were detected and verified through quantitative RT-PCR, and a novel circRNA was screened, which was circLPAR3. Circular RNA LPAR3 showed apparently high expression in ESCC tissues and cells, which was closely correlated with the clinical stage and lymph node metastasis of ESCC patients. Circular RNA LPAR3 was mainly located in the cytoplasm of ESCC cells, which was more stable than the baseline gene. Circular RNA LPAR3 upregulated MET gene expression through sponge adsorption of microRNA (miR)-198, activated the RAS/MAPK and the PI3K/Akt pathways, and promoted ESCC cell migration, invasion, and metastasis in vivo and in vitro. However, it had no effect on ESCC cell proliferation. Circular RNA LPAR3 can regulate the miR-198-MET signal axis to promote the migration, invasion, and metastasis of esophageal cancer cells, which can thereby serve as a potential diagnostic and therapeutic target of esophageal cancer.
Keywords: MET; circLPAR3; circular RNA; esophageal squamous cell carcinoma; miR-198.
© 2020 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Authors declare no conflicts of interest for this article.
Figures




References
-
- Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400‐412. - PubMed
-
- Headrick JR, Nichols FC, Miller DL, et al. High‐grade esophageal dysplasia: long‐term survival and quality of life after esophagectomy. Ann Thorac Surg. 2002;73:1697‐1703. - PubMed
-
- Ohashi S, Miyamoto S, Kikuchi O, et al. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700‐1715. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

