STAT3-mediated MLST8 gene expression regulates cap-dependent translation in cancer cells
- PMID: 32495998
- PMCID: PMC7400782
- DOI: 10.1002/1878-0261.12735
STAT3-mediated MLST8 gene expression regulates cap-dependent translation in cancer cells
Abstract
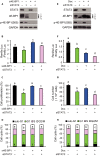
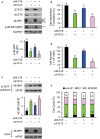
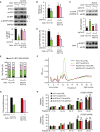
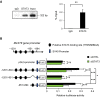
Signal transducer and activator of transcription 3 (STAT3) regulates cell growth, cell survival, angiogenesis, metastasis of cancer cells, and cancer immune evasion by regulating gene expression as a transcription factor. However, the effect of STAT3 on translation is almost unknown. We demonstrated that STAT3 acts as a trans-acting factor for MLST8 gene expression and the protein level of mLST8, a core component of mechanistic target of rapamycin complex 1 and 2 (mTORC1/2), positively regulates the mTORC1/2 downstream pathways. Suppression of STAT3 by siRNA attenuated 4E-BP1 phosphorylation, cap-dependent translation, and cell proliferation in a variety of cancer cells. In HCT116 cells, STAT3 knockdown-induced decreases in 4E-BP1 and AKT phosphorylation levels were further attenuated by MLST8 knockdown or recovered by mLST8 overexpression. STAT3 knockdown-induced G2/M phase arrest was partially restored by co-knockdown of 4EBP1, and the attenuation of cell proliferation was enhanced by the expression of an mTORC1-mediated phosphorylation-defective mutant of 4E-BP1. ChIP and promoter mapping using a luciferase reporter assay showed that the -951 to -894 bp of MLST8 promoter seems to include STAT3-binding site. Overall, these results suggest that STAT3-driven MLST8 gene expression regulates cap-dependent translation through 4E-BP1 phosphorylation in cancer cells.
Keywords: 4E-BP1; MLST8; STAT3; cross-talk; mTORC1.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Disruption of the Scaffolding Function of mLST8 Selectively Inhibits mTORC2 Assembly and Function and Suppresses mTORC2-Dependent Tumor Growth In Vivo.Cancer Res. 2019 Jul 1;79(13):3178-3184. doi: 10.1158/0008-5472.CAN-18-3658. Epub 2019 May 13. Cancer Res. 2019. PMID: 31085701 Free PMC article.
-
Eukaryotic Initiation Factor 4E phosphorylation acts a switch for its binding to 4E-BP1 and mRNA cap assembly.Biochem Biophys Res Commun. 2020 Jun 25;527(2):489-495. doi: 10.1016/j.bbrc.2020.04.086. Epub 2020 Apr 23. Biochem Biophys Res Commun. 2020. PMID: 32336547
-
mLST8 Promotes mTOR-Mediated Tumor Progression.PLoS One. 2015 Apr 23;10(4):e0119015. doi: 10.1371/journal.pone.0119015. eCollection 2015. PLoS One. 2015. PMID: 25906254 Free PMC article.
-
4E-BP1, a multifactor regulated multifunctional protein.Cell Cycle. 2016;15(6):781-6. doi: 10.1080/15384101.2016.1151581. Cell Cycle. 2016. PMID: 26901143 Free PMC article. Review.
-
Reappraisal to the study of 4E-BP1 as an mTOR substrate - A normative critique.Eur J Cell Biol. 2017 Jun;96(4):325-336. doi: 10.1016/j.ejcb.2017.03.013. Epub 2017 Apr 8. Eur J Cell Biol. 2017. PMID: 28427795 Review.
Cited by
-
The Significance of STAT3 in Colonic Diseases: A Comprehensive Study of Pathological Roles and Therapeutic Implications.Cell Biochem Biophys. 2025 Jul 8. doi: 10.1007/s12013-025-01816-0. Online ahead of print. Cell Biochem Biophys. 2025. PMID: 40627320 Review.
-
STAT3 signaling mediates peritoneal fibrosis by activating hyperglycolysis.Am J Transl Res. 2022 Oct 15;14(10):7552-7565. eCollection 2022. Am J Transl Res. 2022. PMID: 36398234 Free PMC article.
-
YAP1 induces bladder cancer progression and promotes immune evasion through IL-6/STAT3 pathway and CXCL deregulation.J Clin Invest. 2024 Dec 4;135(2):e171164. doi: 10.1172/JCI171164. J Clin Invest. 2024. PMID: 39630608 Free PMC article.
-
The mTORC2 signaling network: targets and cross-talks.Biochem J. 2024 Jan 25;481(2):45-91. doi: 10.1042/BCJ20220325. Biochem J. 2024. PMID: 38270460 Free PMC article. Review.
-
Unmasking the tumourigenic role of SIN1/MAPKAP1 in the mTOR complex 2.Clin Transl Med. 2023 Oct;13(10):e1464. doi: 10.1002/ctm2.1464. Clin Transl Med. 2023. PMID: 37877351 Free PMC article. Review.
References
-
- Balendran A, Currie R, Armstrong CG, Avruch J and Alessi DR (1999) Evidence that 3‐phosphoinositide‐dependent protein kinase‐1 mediates phosphorylation of p70 S6 kinase in vivo at Thr‐412 as well as Thr‐252. J Biol Chem 274, 37400–37406. - PubMed
-
- Blenis J (2017) TOR, the gateway to cellular metabolism, cell growth, and disease. Cell 171, 10–13. - PubMed
-
- Cao HH, Tse AK, Kwan HY, Yu H, Cheng CY, Su T, Fong WF and Yu ZL (2014) Quercetin exerts anti‐melanoma activities and inhibits STAT3 signaling. Biochem Pharmacol 87, 424–434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

