A universal method for the rapid isolation of all known classes of functional silencing small RNAs
- PMID: 32496553
- PMCID: PMC7641303
- DOI: 10.1093/nar/gkaa472
A universal method for the rapid isolation of all known classes of functional silencing small RNAs
Abstract
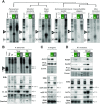
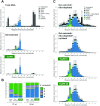
Diverse classes of silencing small (s)RNAs operate via ARGONAUTE-family proteins within RNA-induced-silencing-complexes (RISCs). Here, we have streamlined various embodiments of a Q-sepharose-based RISC-purification method that relies on conserved biochemical properties of all ARGONAUTEs. We show, in multiple benchmarking assays, that the resulting 15-min benchtop extraction procedure allows simultaneous purification of all known classes of RISC-associated sRNAs without prior knowledge of the samples-intrinsic ARGONAUTE repertoires. Optimized under a user-friendly format, the method - coined 'TraPR' for Trans-kingdom, rapid, affordable Purification of RISCs - operates irrespectively of the organism, tissue, cell type or bio-fluid of interest, and scales to minute amounts of input material. The method is highly suited for direct profiling of silencing sRNAs, with TraPR-generated sequencing libraries outperforming those obtained via gold-standard procedures that require immunoprecipitations and/or lengthy polyacrylamide gel-selection. TraPR considerably improves the quality and consistency of silencing sRNA sample preparation including from notoriously difficult-to-handle tissues/bio-fluids such as starchy storage roots or mammalian plasma, and regardless of RNA contaminants or RNA degradation status of samples.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Chalker D.L., Yao M.C.. DNA elimination in ciliates: transposon domestication and genome surveillance. Annu. Rev. Genet. 2011; 45:227–246. - PubMed
-
- Matzke M.A., Mosher R.A.. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014; 15:394–408. - PubMed
-
- McGinn J., Czech B.. Small RNA library construction for high-throughput sequencing. Methods Mol. Biol. 2014; 1093:195–208. - PubMed

