Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors
- PMID: 32496587
- PMCID: PMC7300910
- DOI: 10.1111/all.14429
Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors
Abstract
Background: Morbidity and mortality from COVID-19 caused by novel coronavirus SARS-CoV-2 is accelerating worldwide, and novel clinical presentations of COVID-19 are often reported. The range of human cells and tissues targeted by SARS-CoV-2, its potential receptors and associated regulating factors are still largely unknown. The aim of our study was to analyze the expression of known and potential SARS-CoV-2 receptors and related molecules in the extensive collection of primary human cells and tissues from healthy subjects of different age and from patients with risk factors and known comorbidities of COVID-19.
Methods: We performed RNA sequencing and explored available RNA-Seq databases to study gene expression and co-expression of ACE2, CD147 (BSG), and CD26 (DPP4) and their direct and indirect molecular partners in primary human bronchial epithelial cells, bronchial and skin biopsies, bronchoalveolar lavage fluid, whole blood, peripheral blood mononuclear cells (PBMCs), monocytes, neutrophils, DCs, NK cells, ILC1, ILC2, ILC3, CD4+ and CD8+ T cells, B cells, and plasmablasts. We analyzed the material from healthy children and adults, and from adults in relation to their disease or COVID-19 risk factor status.
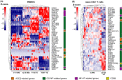
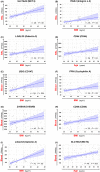
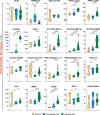
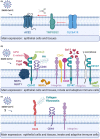
Results: ACE2 and TMPRSS2 were coexpressed at the epithelial sites of the lung and skin, whereas CD147 (BSG), cyclophilins (PPIA andPPIB), CD26 (DPP4), and related molecules were expressed in both epithelium and in immune cells. We also observed a distinct age-related expression profile of these genes in the PBMCs and T cells from healthy children and adults. Asthma, COPD, hypertension, smoking, obesity, and male gender status generally led to the higher expression of ACE2- and CD147-related genes in the bronchial biopsy, BAL, or blood. Additionally, CD147-related genes correlated positively with age and BMI. Interestingly, we also observed higher expression of CD147-related genes in the lesional skin of patients with atopic dermatitis.
Conclusions: Our data suggest different receptor repertoire potentially involved in the SARS-CoV-2 infection at the epithelial barriers and in the immune cells. Altered expression of these receptors related to age, gender, obesity and smoking, as well as with the disease status, might contribute to COVID-19 morbidity and severity patterns.
Keywords: COPD; COVID-19; COVID-19 children; SARS receptor; asthma; hypertension; obesity.
© 2020 EAACI and John Wiley and Sons A/S. Published by John Wiley and Sons Ltd.
Conflict of interest statement
Dr Radzikowska has nothing to disclose. Dr Ding has nothing to disclose. Dr Tan has nothing to disclose. Dr Zhakparov has nothing to disclose. Dr Peng has nothing to disclose. Dr Wawrzyniak has nothing to disclose. Dr Wang has nothing to disclose. Dr LI has nothing to disclose. Dr Morita has nothing to disclose. Dr Altunbulakli has nothing to disclose. Dr Reiger reports personal fees from Bencard, Germany, personal fees from Roche‐Posay, Germany, personal fees from Galderma, Germany, personal fees from Sebapharma, Germany, grants from CLR, Germany, outside the submitted work. Dr Neumann has nothing to disclose. Dr Lunjani has nothing to disclose. Dr Traidl‐Hoffmann reports grants and personal fees from Töpfer, personal fees from Sanofi, personal fees from Novartis, grants and personal fees from Sebapharma, grants and personal fees from Danone Nutricia, personal fees from La Roche Posay, personal fees from Lilly Pharma, personal fees from Mylan, outside the submitted work. Dr Nadeau reports grants and other from NIAID, other from Novartis, personal fees and other from Regeneron, grants and other from FARE, grants from EAT, other from Sanofi, other from Astellas, other from Nestle, other from BeforeBrands, other from Alladapt, other from ForTra, other from Genentech, other from AImmune Therapeutics, other from DBV Technologies, personal fees from Astrazeneca, personal fees from ImmuneWorks, personal fees from Cour Pharmaceuticals, grants from Allergenis, grants from Ukko Pharma, other from AnaptysBio, other from Adare Pharmaceuticals, other from Stallergenes‐Greer, other from NHLBI, other from NIEHS, other from EPA, other from WAO Center of Excellence, other from Iggenix, other from Probio, other from Vedanta, other from Centecor, other from Seed, from Immune Tolerance Network, from NIH, outside the submitted work. In addition, Dr Nadeau has a patent Inhibition of Allergic Reaction to Peanut Allergen using an IL‐33 Inhibitor pending, a patent Special Oral Formula for Decreasing Food Allergy Risk and Treatment for Food Allergy pending, a patent Basophil Activation Based Diagnostic Allergy Test pending, a patent Granulocyte‐based methods for detecting and monitoring immune system disorders pending, a patent Methods and Assays for Detecting and Quantifying Pure Subpopulations of White Blood Cells in Immune System Disorders pending, a patent Mixed Allergen Compositions and Methods for Using the Same pending, and a patent Microfluidic Device and Diagnostic Methods for Allergy Testing Based on Detection of Basophil Activation pending. Dr O'Mahony reports personal fees from AHL, grants from GSK, outside the submitted work. Dr Akdis reports grants from Allergopharma, Idorsia, Swiss National Science Foundation, Christine Kühne‐Center for Allergy Research and Education, European Commission's Horison's 2020 Framework Programme, Cure, Novartis Research Institutes, Astra Zeneca, Scibase, Glakso Smith‐Kline and other from Sanofi & Regeneron. Dr Sokolowska reports grants from SNSF, grants from GSK, outside the submitted work.
Figures


Comment in
-
CD147 as a novel receptor in the pathogenesis of SARS-CoV-2: Is there any correlation with the risk of COVID-19 in dermatological diseases?Dermatol Ther. 2020 Nov;33(6):e14443. doi: 10.1111/dth.14443. Epub 2020 Nov 15. Dermatol Ther. 2020. PMID: 33089902 Free PMC article. No abstract available.
References
-
- Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy 2020;75:1730‐1741. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

