Developmental alterations in the transcriptome of three distinct rodent models of schizophrenia
- PMID: 32497066
- PMCID: PMC7272013
- DOI: 10.1371/journal.pone.0232200
Developmental alterations in the transcriptome of three distinct rodent models of schizophrenia
Abstract
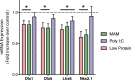
Schizophrenia is a debilitating disorder affecting just under 1% of the population. While the symptoms of this disorder do not appear until late adolescence, pathological alterations likely occur earlier, during development in utero. While there is an increasing literature examining transcriptome alterations in patients, it is not possible to examine the changes in gene expression that occur during development in humans that will develop schizophrenia. Here we utilize three distinct rodent developmental disruption models of schizophrenia to examine potential overlapping alterations in the transcriptome, with a specific focus on markers of interneuron development. Specifically, we administered either methylazoxymethanol acetate (MAM), Polyinosinic:polycytidylic acid (Poly I:C), or chronic protein malnutrition, on GD 17 and examined mRNA expression in the developing hippocampus of the offspring 18 hours later. Here, we report alterations in gene expression that may contribute to the pathophysiology of schizophrenia, including significant alterations in interneuron development and ribosome function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

