Identification of Clec4b as a novel regulator of bystander activation of auto-reactive T cells and autoimmune disease
- PMID: 32497089
- PMCID: PMC7297379
- DOI: 10.1371/journal.pgen.1008788
Identification of Clec4b as a novel regulator of bystander activation of auto-reactive T cells and autoimmune disease
Abstract
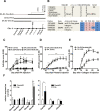
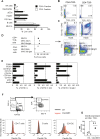
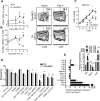
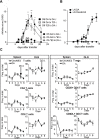
The control of chronic inflammation is dependent on the possibility of limiting bystander activation of autoreactive and potentially pathogenic T cells. We have identified a non-sense loss of function single nucleotide polymorphism in the C-type lectin receptor, Clec4b, and have shown that it controls chronic autoimmune arthritis in rat models of rheumatoid arthritis. Clec4b is specifically expressed in CD4+ myeloid cells, mainly classical dendritic cells (DCs), and is defined by the markers CD4+/MHCIIhi/CD11b/c+. We found that Clec4b limited the activation of arthritogenic CD4+αβT cells and the absence of Clec4b allowed development of arthritis already 5 days after adjuvant injection. Clec4b sufficient CD4+ myeloid dendritic cells successfully limited the arthritogenic T cell expansion immediately after activation both in vitro and in vivo. We conclude that Clec4b expressed on CD4+ myeloid dendritic cells regulate the expansion of auto-reactive and potentially pathogenic T cells during an immune response, demonstrating an early checkpoint control mechanism to avoid autoimmunity leading to chronic inflammation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Vingsbo C, Sahlstrand P, Brun JG, Jonsson R, Saxne T, Holmdahl R. Pristane-induced arthritis in rats: a new model for rheumatoid arthritis with a chronic disease course influenced by both major histocompatibility complex and non-major histocompatibility complex genes. Am J Pathol. American Society for Investigative Pathology; 1996. November;149(5):1675–83. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

