Mechanistic dissection of the PD-L1:B7-1 co-inhibitory immune complex
- PMID: 32497097
- PMCID: PMC7272049
- DOI: 10.1371/journal.pone.0233578
Mechanistic dissection of the PD-L1:B7-1 co-inhibitory immune complex
Abstract
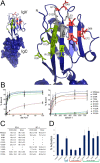
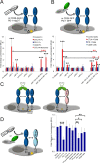
The B7 family represents one of the best-studied subgroups within the Ig superfamily, yet new interactions continue to be discovered. However, this binding promiscuity represents a major challenge for defining the biological contribution of each specific interaction. We developed a strategy for addressing these challenges by combining cell microarray and high-throughput FACS methods to screen for promiscuous binding events, map binding interfaces, and generate functionally selective reagents. Applying this approach to the interactions of mPD-L1 with its receptor mPD-1 and its ligand mB7-1, we identified the binding interface of mB7-1 on mPD-L1 and as a result generated mPD-L1 mutants with binding selectivity for mB7-1 or mPD-1. Next, using a panel of mB7-1 mutants, we mapped the binding sites of mCTLA-4, mCD28 and mPD-L1. Surprisingly, the mPD-L1 binding site mapped to the dimer interface surface of mB7-1, placing it distal from the CTLA-4/CD28 recognition surface. Using two independent approaches, we demonstrated that mPD-L1 and mB7-1 bind in cis, consistent with recent reports from Chaudhri A et al. and Sugiura D et al. We further provide evidence that while CTLA-4 and CD28 do not directly compete with PD-L1 for binding to B7-1, they can disrupt the cis PD-L1:B7-1 complex by reorganizing B7-1 on the cell surface. These observations offer new functional insights into the regulatory mechanisms associated with this group of B7 family proteins and provide new tools to elucidate their function in vitro and in vivo.
Conflict of interest statement
We acknowledge a relationship to Cue Biopharma, Inc. Technologies described in this manuscript were disclosed in PCT patent application nos. PCT/US2013/073275,PCT/US2015/035777, and PCT/US2017/33042, and their corresponding national and regional patents and patent applications, all of which are licensed to Cue Biopharma, Inc. Almo holds equity in Cue Biopharma, Inc. and is a member of its Scientific Advisory Board. However, this commercial affiliation does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures






References
-
- Wang S, Chen L (2004) T lymphocyte co-signaling pathways of the B7-CD28 family. Cell Mol Immunol 1: 37–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

