Increasing host cellular receptor-angiotensin-converting enzyme 2 expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV-2) infection
- PMID: 32497323
- PMCID: PMC7300907
- DOI: 10.1002/jmv.26139
Increasing host cellular receptor-angiotensin-converting enzyme 2 expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV-2) infection
Abstract
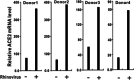
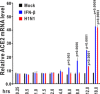
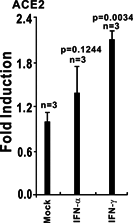
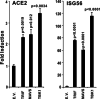
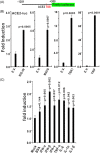
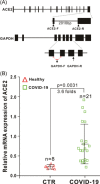
The ongoing outbreak of a new coronavirus (2019-nCoV, or severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]) has caused an epidemic of the acute respiratory syndrome known as coronavirus disease (COVID-19) in humans. SARS-CoV-2 rapidly spread to multiple regions of China and multiple other countries, posing a serious threat to public health. The spike (S) proteins of SARS-CoV-1 and SARS-CoV-2 may use the same host cellular receptor, angiotensin-converting enzyme 2 (ACE2), for entering host cells. The affinity between ACE2 and the SARS-CoV-2 S protein is much higher than that of ACE2 binding to the SARS-CoV S protein, explaining why SARS-CoV-2 seems to be more readily transmitted from human to human. Here, we report that ACE2 can be significantly upregulated after infection of various viruses, including SARS-CoV-1 and SARS-CoV-2, or by the stimulation with inflammatory cytokines such as interferons. We propose that SARS-CoV-2 may positively induce its cellular entry receptor, ACE2, to accelerate its replication and spread; high inflammatory cytokine levels increase ACE2 expression and act as high-risk factors for developing COVID-19, and the infection of other viruses may increase the risk of SARS-CoV-2 infection. Therefore, drugs targeting ACE2 may be developed for the future emerging infectious diseases caused by this cluster of coronaviruses.
Keywords: 2019-nCoV; ACE2; COVID-19; IFN; ISG; SARS-CoV-2.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

