Hydrophobic Amino Acids as Universal Elements of Protein-Induced DNA Structure Deformation
- PMID: 32498246
- PMCID: PMC7312683
- DOI: 10.3390/ijms21113986
Hydrophobic Amino Acids as Universal Elements of Protein-Induced DNA Structure Deformation
Abstract
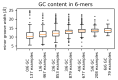
Interaction with the DNA minor groove is a significant contributor to specific sequence recognition in selected families of DNA-binding proteins. Based on a statistical analysis of 3D structures of protein-DNA complexes, we propose that distortion of the DNA minor groove resulting from interactions with hydrophobic amino acid residues is a universal element of protein-DNA recognition. We provide evidence to support this by associating each DNA minor groove-binding amino acid residue with the local dimensions of the DNA double helix using a novel algorithm. The widened DNA minor grooves are associated with high GC content. However, some AT-rich sequences contacted by hydrophobic amino acids (e.g., phenylalanine) display extreme values of minor groove width as well. For a number of hydrophobic amino acids, distinct secondary structure preferences could be identified for residues interacting with the widened DNA minor groove. These results hold even after discarding the most populous families of minor groove-binding proteins.
Keywords: DNA shape; hydrophobic; indirect readout; minor groove; protein–DNA interaction; specific recognition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


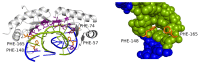


Similar articles
-
Statistical analysis of structural determinants for protein-DNA-binding specificity.Proteins. 2016 Aug;84(8):1147-61. doi: 10.1002/prot.25061. Epub 2016 Jun 15. Proteins. 2016. PMID: 27147539 Free PMC article.
-
Protein-DNA hydrophobic recognition in the minor groove is facilitated by sugar switching.J Mol Biol. 2004 Mar 12;337(1):65-76. doi: 10.1016/j.jmb.2004.01.011. J Mol Biol. 2004. PMID: 15001352
-
Solution structure of the calicheamicin gamma 1I-DNA complex.J Mol Biol. 1997 Jan 17;265(2):187-201. doi: 10.1006/jmbi.1996.0718. J Mol Biol. 1997. PMID: 9020982
-
What drives proteins into the major or minor grooves of DNA?J Mol Biol. 2007 Jan 5;365(1):1-9. doi: 10.1016/j.jmb.2006.09.059. Epub 2006 Sep 27. J Mol Biol. 2007. PMID: 17055530 Free PMC article. Review.
-
DNA recognition by beta-sheets.Biopolymers. 1997;44(4):335-59. doi: 10.1002/(SICI)1097-0282(1997)44:4<335::AID-BIP3>3.0.CO;2-R. Biopolymers. 1997. PMID: 9782775 Review.
Cited by
-
Cryo-EM Structure of AAV2 Rep68 bound to integration site AAVS1: Insights into the mechanism of DNA melting.bioRxiv [Preprint]. 2024 Apr 2:2024.04.02.587759. doi: 10.1101/2024.04.02.587759. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Jan 24;53(3):gkaf033. doi: 10.1093/nar/gkaf033. PMID: 38617369 Free PMC article. Updated. Preprint.
-
Chia Derived Peptides Affecting Bacterial Membrane and DNA: Insights from Staphylococcus aureus and Escherichia coli Studies.Plant Foods Hum Nutr. 2024 Dec 28;80(1):22. doi: 10.1007/s11130-024-01240-4. Plant Foods Hum Nutr. 2024. PMID: 39733059
-
Identification and Functional Analysis of a Novel Hydrophobic Protein VdHP1 from Verticillium dahliae.Microbiol Spectr. 2022 Apr 27;10(2):e0247821. doi: 10.1128/spectrum.02478-21. Epub 2022 Apr 4. Microbiol Spectr. 2022. PMID: 35377232 Free PMC article.
-
Cryo-EM structure of AAV2 Rep68 bound to integration site AAVS1: insights into the mechanism of DNA melting.Nucleic Acids Res. 2025 Jan 24;53(3):gkaf033. doi: 10.1093/nar/gkaf033. Nucleic Acids Res. 2025. PMID: 39883011 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

