Skeletal Muscle Gene Expression in Long-Term Endurance and Resistance Trained Elderly
- PMID: 32498275
- PMCID: PMC7312229
- DOI: 10.3390/ijms21113988
Skeletal Muscle Gene Expression in Long-Term Endurance and Resistance Trained Elderly
Abstract
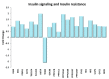
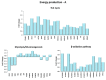
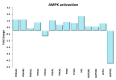
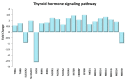
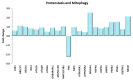
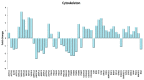
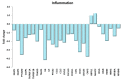
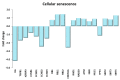
Physical exercise is deemed the most efficient way of counteracting the age-related decline of skeletal muscle. Here we report a transcriptional study by next-generation sequencing of vastus lateralis biopsies from elderly with a life-long high-level training practice (n = 9) and from age-matched sedentary subjects (n = 5). Unsupervised mixture distribution analysis was able to correctly categorize trained and untrained subjects, whereas it failed to discriminate between individuals who underwent a prevalent endurance (n = 5) or a prevalent resistance (n = 4) training, thus showing that the training mode was not relevant for sarcopenia prevention. KEGG analysis of transcripts showed that physical exercise affected a high number of metabolic and signaling pathways, in particular those related to energy handling and mitochondrial biogenesis, where AMPK and AKT-mTOR signaling pathways are both active and balance each other, concurring to the establishment of an insulin-sensitive phenotype and to the maintenance of a functional muscle mass. Other pathways affected by exercise training increased the efficiency of the proteostatic mechanisms, consolidated the cytoskeletal organization, lowered the inflammation level, and contrasted cellular senescence. This study on extraordinary individuals who trained at high level for at least thirty years suggests that aging processes and exercise training travel the same paths in the opposite direction.
Keywords: aging; endurance and resistance training; exercise; gene expression; sarcopenia; skeletal muscle.
Conflict of interest statement
The Authors declare the absence of conflicts of interest.
Figures






Similar articles
-
Non-Coding RNAs in the Transcriptional Network That Differentiates Skeletal Muscles of Sedentary from Long-Term Endurance- and Resistance-Trained Elderly.Int J Mol Sci. 2021 Feb 3;22(4):1539. doi: 10.3390/ijms22041539. Int J Mol Sci. 2021. PMID: 33546468 Free PMC article.
-
Effects of Training Status and Exercise Mode on Global Gene Expression in Skeletal Muscle.Int J Mol Sci. 2021 Nov 22;22(22):12578. doi: 10.3390/ijms222212578. Int J Mol Sci. 2021. PMID: 34830458 Free PMC article.
-
Resistance exercise enhances the molecular signaling of mitochondrial biogenesis induced by endurance exercise in human skeletal muscle.J Appl Physiol (1985). 2011 Nov;111(5):1335-44. doi: 10.1152/japplphysiol.00086.2011. Epub 2011 Aug 11. J Appl Physiol (1985). 2011. PMID: 21836044 Clinical Trial.
-
Manipulating Cellular Energetics to Slow Aging of Tissues and Organs.Biochemistry (Mosc). 2020 Jun;85(6):651-659. doi: 10.1134/S0006297920060024. Biochemistry (Mosc). 2020. PMID: 32586228 Review.
-
Protein intake and amino acid supplementation regulate exercise recovery and performance through the modulation of mTOR, AMPK, FGF21, and immunity.Nutr Res. 2019 Dec;72:1-17. doi: 10.1016/j.nutres.2019.06.006. Epub 2019 Jul 2. Nutr Res. 2019. PMID: 31672317 Review.
Cited by
-
Regulatory Mechanisms of Yili Horses During an 80 km Race Based on Transcriptomics and Metabolomics Analyses.Int J Mol Sci. 2025 Mar 8;26(6):2426. doi: 10.3390/ijms26062426. Int J Mol Sci. 2025. PMID: 40141070 Free PMC article.
-
The relationship between sarcopenia and related bioindicators and changes after intensive lifestyle intervention in elderly East-China populations.BMC Musculoskelet Disord. 2024 Sep 3;25(1):704. doi: 10.1186/s12891-024-07835-x. BMC Musculoskelet Disord. 2024. PMID: 39227842 Free PMC article. Clinical Trial.
-
Physical activity and exercise in the context of SARS-Cov-2: A perspective from geroscience field.Ageing Res Rev. 2021 Mar;66:101258. doi: 10.1016/j.arr.2021.101258. Epub 2021 Jan 12. Ageing Res Rev. 2021. PMID: 33450400 Free PMC article. Review.
-
Genome-Wide Association Study of Exercise-Induced Fat Loss Efficiency.Genes (Basel). 2022 Oct 29;13(11):1975. doi: 10.3390/genes13111975. Genes (Basel). 2022. PMID: 36360211 Free PMC article.
-
De novo Explorations of Sarcopenia via a Dynamic Model.Front Physiol. 2021 May 28;12:670381. doi: 10.3389/fphys.2021.670381. eCollection 2021. Front Physiol. 2021. PMID: 34122142 Free PMC article.
References
-
- Hwang A.B., Brack A.S. Muscle Stem Cells and Aging. Curr. Top. Dev. Biol. 2018;126:299–322. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

