Detection and Characterization of Porcine Sapelovirus in Italian Pig Farms
- PMID: 32498384
- PMCID: PMC7341194
- DOI: 10.3390/ani10060966
Detection and Characterization of Porcine Sapelovirus in Italian Pig Farms
Abstract
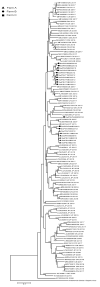
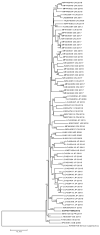
Porcine sapelovirus (PSV) belongs to the genus Sapelovirus of the family Picornaviridae. PSV infects pigs asymptomatically, but it can also cause severe neurologic, enteric, and respiratory symptoms or reproductive failure. Sapelovirus infections have been reported worldwide in pigs. The objective of this study was to investigate the presence and the prevalence of PSV in Italian swine farms in animals of different ages to clarify the occurrence of the infection and the genetic characteristics of circulating strains. In the present study, 92 pools of fecal samples, collected from pigs across three farms, were analyzed by Reverse Transcriptase-polymerase Chain Reaction-PCR (RT-PCR). Fecal pools from young growers (63/64) were found positive for Sapelovirus in all farms while detection in sows (4/28) was observed in only one farm. Phylogenetic analyses of the 19 partial capsid protein nucleotide sequences (VP1) (6-7 each farm) enable the classification of the virus sequences into three distinct clades and highlighted the high heterogeneity within one farm. The whole genome sequence obtained from one strain showed the highest correlation with the Italian strain detected in 2015. The study adds novel information about the circulation and heterogeneity of PSV strains in Italy and considering the movement of pigs across Europe would also be informative for other countries.
Keywords: Italy; PSV; porcine sapelovirus; swine.
Conflict of interest statement
None of the authors has any other financial or personal relationships that could inappropriately influence or bias the content of the paper.
Figures
Similar articles
-
High prevalence, genetic diversity and a potentially novel genotype of Sapelovirus A (Picornaviridae) in enteric and respiratory samples in Hungarian swine farms.J Gen Virol. 2020 Jun;101(6):609-621. doi: 10.1099/jgv.0.001410. Epub 2020 Apr 7. J Gen Virol. 2020. PMID: 32255421 Free PMC article.
-
Characterization of porcine sapelovirus prevalent in western Jiangxi, China.BMC Vet Res. 2021 Aug 14;17(1):273. doi: 10.1186/s12917-021-02979-7. BMC Vet Res. 2021. PMID: 34391425 Free PMC article.
-
Porcine sapelovirus among diarrhoeic piglets in India.Transbound Emerg Dis. 2018 Feb;65(1):261-263. doi: 10.1111/tbed.12628. Epub 2017 Feb 27. Transbound Emerg Dis. 2018. PMID: 28244229
-
High Prevalence, Genetic Diversity, and Recombination of Porcine Sapelovirus in Pig Farms in Fujian, Southern China.Viruses. 2023 Aug 17;15(8):1751. doi: 10.3390/v15081751. Viruses. 2023. PMID: 37632093 Free PMC article.
-
First report of Porcine teschovirus (PTV), Porcine sapelovirus (PSV) and Enterovirus G (EV-G) in pig herds of Brazil.Trop Anim Health Prod. 2014 Mar;46(3):523-8. doi: 10.1007/s11250-013-0523-z. Epub 2013 Dec 22. Trop Anim Health Prod. 2014. PMID: 24362793
Cited by
-
Isolation, Characterization, and Molecular Detection of Porcine Sapelovirus.Viruses. 2022 Feb 8;14(2):349. doi: 10.3390/v14020349. Viruses. 2022. PMID: 35215935 Free PMC article.
-
Relevancy Prediction of the Emerging Pathogens with Porcine Diarrhea by Logistic Regression Model.Microorganisms. 2025 Feb 27;13(3):528. doi: 10.3390/microorganisms13030528. Microorganisms. 2025. PMID: 40142424 Free PMC article.
-
Exploring the Cause of Diarrhoea and Poor Growth in 8-11-Week-Old Pigs from an Australian Pig Herd Using Metagenomic Sequencing.Viruses. 2021 Aug 13;13(8):1608. doi: 10.3390/v13081608. Viruses. 2021. PMID: 34452472 Free PMC article.
-
Porcine teschovirus, sapelovirus, and enterovirus in Swiss pigs: multiplex RT-PCR investigation of viral frequencies and disease association.J Vet Diagn Invest. 2021 Sep;33(5):864-874. doi: 10.1177/10406387211025827. Epub 2021 Jun 21. J Vet Diagn Invest. 2021. PMID: 34151653 Free PMC article.
References
-
- Adams M.J., Lefkowitz E.J., King A.M.Q., Bamford D., Breitbart M., Davison A.J., Ghabrial S.A., Gorbalenya A.E., Knowles N.J., Krell P., et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2015) Arch. Virol. 2015;160:1837–1850. doi: 10.1007/s00705-015-2425-z. - DOI - PubMed
LinkOut - more resources
Full Text Sources

