Bayesian and Machine Learning Models for Genomic Prediction of Anterior Cruciate Ligament Rupture in the Canine Model
- PMID: 32499222
- PMCID: PMC7407450
- DOI: 10.1534/g3.120.401244
Bayesian and Machine Learning Models for Genomic Prediction of Anterior Cruciate Ligament Rupture in the Canine Model
Abstract
Anterior cruciate ligament (ACL) rupture is a common, debilitating condition that leads to early-onset osteoarthritis and reduced quality of human life. ACL rupture is a complex disease with both genetic and environmental risk factors. Characterizing the genetic basis of ACL rupture would provide the ability to identify individuals that have high genetic risk and allow the opportunity for preventative management. Spontaneous ACL rupture is also common in dogs and shows a similar clinical presentation and progression. Thus, the dog has emerged as an excellent genomic model for human ACL rupture. Genome-wide association studies (GWAS) in the dog have identified a number of candidate genetic variants, but research in genomic prediction has been limited. In this analysis, we explore several Bayesian and machine learning models for genomic prediction of ACL rupture in the Labrador Retriever dog. Our work demonstrates the feasibility of predicting ACL rupture from SNPs in the Labrador Retriever model with and without consideration of non-genetic risk factors. Genomic prediction including non-genetic risk factors approached clinical relevance using multiple linear Bayesian and non-linear models. This analysis represents the first steps toward development of a predictive algorithm for ACL rupture in the Labrador Retriever model. Future work may extend this algorithm to other high-risk breeds of dog. The ability to accurately predict individual dogs at high risk for ACL rupture would identify candidates for clinical trials that would benefit both veterinary and human medicine.
Keywords: ACL rupture; Canine; Dog model; GenPred; Genomic prediction; Shared data resources.
Copyright © 2020 Baker et al.
Figures
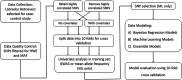
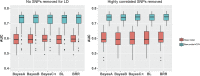
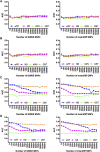
References
-
- Albert J. H., and Chib S., 1993. Bayesian analysis of binary and polychotomous response data. J. Am. Stat. Assoc. 88: 669–679. 10.1080/01621459.1993.10476321 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
