First-in-Class Inhibitors of Oncogenic CHD1L with Preclinical Activity against Colorectal Cancer
- PMID: 32499299
- PMCID: PMC7665848
- DOI: 10.1158/1535-7163.MCT-20-0106
First-in-Class Inhibitors of Oncogenic CHD1L with Preclinical Activity against Colorectal Cancer
Abstract
Since the discovery of CHD1L in 2008, it has emerged as an oncogene implicated in the pathology and poor prognosis of a variety of cancers, including gastrointestinal cancers. However, a mechanistic understanding of CHD1L as a driver of colorectal cancer has been limited. Until now, there have been no reported inhibitors of CHD1L, also limiting its development as a molecular target. We sought to characterize the clinicopathologic link between CHD1L and colorectal cancer, determine the mechanism(s) by which CHD1L drives malignant colorectal cancer, and discover the first inhibitors with potential for novel treatments for colorectal cancer. The clinicopathologic characteristics associated with CHD1L expression were evaluated using microarray data from 585 patients with colorectal cancer. Further analysis of microarray data indicated that CHD1L may function through the Wnt/TCF pathway. Thus, we conducted knockdown and overexpression studies with CHD1L to determine its role in Wnt/TCF-driven epithelial-to-mesenchymal transition (EMT). We performed high-throughput screening (HTS) to identify the first CHD1L inhibitors. The mechanism of action, antitumor efficacy, and drug-like properties of lead CHD1L inhibitors were determined using biochemical assays, cell models, tumor organoids, patient-derived tumor organoids, and in vivo pharmacokinetics and pharmacodynamics. Lead CHD1L inhibitors display potent in vitro antitumor activity by reversing TCF-driven EMT. The best lead CHD1L inhibitor possesses drug-like properties in pharmacokinetic/pharmacodynamic mouse models. This work validates CHD1L as a druggable target and establishes a novel therapeutic strategy for the treatment of colorectal cancer.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures

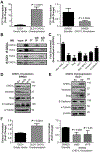
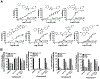
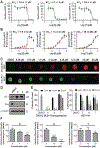

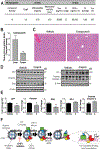
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

