Modular and Distinct Plexin-A4/FARP2/Rac1 Signaling Controls Dendrite Morphogenesis
- PMID: 32499377
- PMCID: PMC7343331
- DOI: 10.1523/JNEUROSCI.2730-19.2020
Modular and Distinct Plexin-A4/FARP2/Rac1 Signaling Controls Dendrite Morphogenesis
Abstract
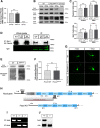
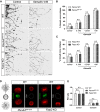
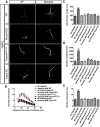
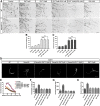
Diverse neuronal populations with distinct cellular morphologies coordinate the complex function of the nervous system. Establishment of distinct neuronal morphologies critically depends on signaling pathways that control axonal and dendritic development. The Sema3A-Nrp1/PlxnA4 signaling pathway promotes cortical neuron basal dendrite arborization but also repels axons. However, the downstream signaling components underlying these disparate functions of Sema3A signaling are unclear. Using the novel PlxnA4KRK-AAA knock-in male and female mice, generated by CRISPR/cas9, we show here that the KRK motif in the PlxnA4 cytoplasmic domain is required for Sema3A-mediated cortical neuron dendritic elaboration but is dispensable for inhibitory axon guidance. The RhoGEF FARP2, which binds to the KRK motif, shows identical functional specificity as the KRK motif in the PlxnA4 receptor. We find that Sema3A activates the small GTPase Rac1, and that Rac1 activity is required for dendrite elaboration but not axon growth cone collapse. This work identifies a novel Sema3A-Nrp1/PlxnA4/FARP2/Rac1 signaling pathway that specifically controls dendritic morphogenesis but is dispensable for repulsive guidance events. Overall, our results demonstrate that the divergent signaling output from multifunctional receptor complexes critically depends on distinct signaling motifs, highlighting the modular nature of guidance cue receptors and its potential to regulate diverse cellular responses.SIGNIFICANCE STATEMENT The proper formation of axonal and dendritic morphologies is crucial for the precise wiring of the nervous system that ultimately leads to the generation of complex functions in an organism. The Semaphorin3A-Neuropilin1/Plexin-A4 signaling pathway has been shown to have multiple key roles in neurodevelopment, from axon repulsion to dendrite elaboration. This study demonstrates that three specific amino acids, the KRK motif within the Plexin-A4 receptor cytoplasmic domain, are required to coordinate the downstream signaling molecules to promote Sema3A-mediated cortical neuron dendritic elaboration, but not inhibitory axon guidance. Our results unravel a novel Semaphorin3A-Plexin-A4 downstream signaling pathway and shed light on how the disparate functions of axon guidance and dendritic morphogenesis are accomplished by the same extracellular ligand in vivo.
Keywords: Semaphorin signaling; axonal repulsion; dendritic branching; guidance receptors; neural development.
Copyright © 2020 the authors.
Figures





Similar articles
-
Balancing act of small GTPases downstream of plexin-A4 signaling motifs promotes dendrite elaboration in mammalian cortical neurons.Sci Signal. 2024 Jan 16;17(819):eadh7673. doi: 10.1126/scisignal.adh7673. Epub 2024 Jan 16. Sci Signal. 2024. PMID: 38227686
-
Distinct cytoplasmic domains in Plexin-A4 mediate diverse responses to semaphorin 3A in developing mammalian neurons.Sci Signal. 2014 Mar 11;7(316):ra24. doi: 10.1126/scisignal.2004734. Sci Signal. 2014. PMID: 24619647
-
FARP2 triggers signals for Sema3A-mediated axonal repulsion.Nat Neurosci. 2005 Dec;8(12):1712-9. doi: 10.1038/nn1596. Epub 2005 Nov 13. Nat Neurosci. 2005. PMID: 16286926
-
Molecular basis of semaphorin-mediated axon guidance.J Neurobiol. 2000 Aug;44(2):219-29. doi: 10.1002/1097-4695(200008)44:2<219::aid-neu11>3.0.co;2-w. J Neurobiol. 2000. PMID: 10934324 Review.
-
Regulation of dendritic development by semaphorin 3A through novel intracellular remote signaling.Cell Adh Migr. 2016 Nov;10(6):627-640. doi: 10.1080/19336918.2016.1210758. Epub 2016 Jul 8. Cell Adh Migr. 2016. PMID: 27392015 Free PMC article. Review.
Cited by
-
Semaphorin signaling restricts neuronal regeneration in C. elegans.Front Cell Dev Biol. 2022 Oct 17;10:814160. doi: 10.3389/fcell.2022.814160. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36325362 Free PMC article.
-
Reduced hippocampal inhibition and enhanced autism-epilepsy comorbidity in mice lacking neuropilin 2.Transl Psychiatry. 2021 Oct 18;11(1):537. doi: 10.1038/s41398-021-01655-6. Transl Psychiatry. 2021. PMID: 34663783 Free PMC article.
-
Modelling and Refining Neuronal Circuits with Guidance Cues: Involvement of Semaphorins.Int J Mol Sci. 2021 Jun 6;22(11):6111. doi: 10.3390/ijms22116111. Int J Mol Sci. 2021. PMID: 34204060 Free PMC article. Review.
-
Palmitoylation regulates neuropilin-2 localization and function in cortical neurons and conveys specificity to semaphorin signaling via palmitoyl acyltransferases.Elife. 2023 Apr 3;12:e83217. doi: 10.7554/eLife.83217. Elife. 2023. PMID: 37010951 Free PMC article.
-
Farnesol brain transcriptomics in CNS inflammatory demyelination.Clin Immunol. 2023 Oct;255:109752. doi: 10.1016/j.clim.2023.109752. Epub 2023 Sep 4. Clin Immunol. 2023. PMID: 37673223 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
