CRISPR screen in regulatory T cells reveals modulators of Foxp3
- PMID: 32499641
- PMCID: PMC7305989
- DOI: 10.1038/s41586-020-2246-4
CRISPR screen in regulatory T cells reveals modulators of Foxp3
Abstract
Regulatory T (Treg) cells are required to control immune responses and maintain homeostasis, but are a significant barrier to antitumour immunity1. Conversely, Treg instability, characterized by loss of the master transcription factor Foxp3 and acquisition of proinflammatory properties2, can promote autoimmunity and/or facilitate more effective tumour immunity3,4. A comprehensive understanding of the pathways that regulate Foxp3 could lead to more effective Treg therapies for autoimmune disease and cancer. The availability of new functional genetic tools has enabled the possibility of systematic dissection of the gene regulatory programs that modulate Foxp3 expression. Here we developed a CRISPR-based pooled screening platform for phenotypes in primary mouse Treg cells and applied this technology to perform a targeted loss-of-function screen of around 500 nuclear factors to identify gene regulatory programs that promote or disrupt Foxp3 expression. We identified several modulators of Foxp3 expression, including ubiquitin-specific peptidase 22 (Usp22) and ring finger protein 20 (Rnf20). Usp22, a member of the deubiquitination module of the SAGA chromatin-modifying complex, was revealed to be a positive regulator that stabilized Foxp3 expression; whereas the screen suggested that Rnf20, an E3 ubiquitin ligase, can serve as a negative regulator of Foxp3. Treg-specific ablation of Usp22 in mice reduced Foxp3 protein levels and caused defects in their suppressive function that led to spontaneous autoimmunity but protected against tumour growth in multiple cancer models. Foxp3 destabilization in Usp22-deficient Treg cells could be rescued by ablation of Rnf20, revealing a reciprocal ubiquitin switch in Treg cells. These results reveal previously unknown modulators of Foxp3 and demonstrate a screening method that can be broadly applied to discover new targets for Treg immunotherapies for cancer and autoimmune disease.
Conflict of interest statement
Declaration of Interests
The authors declare competing financial interests: T.L.R. is a co-founder of Arsenal Biosciences. A.M. is a co-founder of Spotlight Therapeutics and Arsenal Biosciences. A.M. has served as an advisor to Juno Therapeutics, is a member of the scientific advisory board at PACT Pharma, and is an advisor to Trizell. A.M. owns stock in Arsenal Biosciences, Spotlight Therapeutics and PACT Pharma. The Marson lab has received sponsored research support from Juno Therapeutics, Epinomics, Sanofi, and a gift from Gilead. J.A.B. is a co-founder of Sonoma BioTherapeutics; a consultant for Juno, a Celgene company; a stock holder and member of the Board of Directors on Rheos Medicines; and a stock holder and member of the Scientific Advisory Boards of Pfizer Center for Therapeutic Innovation, Vir Therapeutics, Arcus Biotherapeutics, Quentis Therapeutics, Solid Biosciences, and Celsius Therapeutics. J.A.B. owns stock in MacroGenics Inc., Vir Therapeutics, Arcus Biotherapeutics, Quentis Therapeutics, Solid Biosciences, Celsius Therapeutics, and Kadmon Holdings. A patent application has been filed based on the screen data described here.
Figures



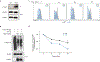





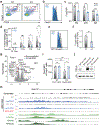
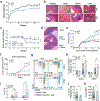
References
Method References
-
- Luche H, Weber O, Nageswara Rao T, Blum C & Fehling HJ Faithful activation of an extra-bright red fluorescent protein in ‘knock-in’ Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol 37, 43–53 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 CA184043/CA/NCI NIH HHS/United States
- P30 DK063720/DK/NIDDK NIH HHS/United States
- T32 GM007618/GM/NIGMS NIH HHS/United States
- R35 GM128943/GM/NIGMS NIH HHS/United States
- T32 CA009370/CA/NCI NIH HHS/United States
- S10 OD021822/OD/NIH HHS/United States
- R01 AI151123/AI/NIAID NIH HHS/United States
- T32 AI125222/AI/NIAID NIH HHS/United States
- T32 GM008568/GM/NIGMS NIH HHS/United States
- T32 AI007334/AI/NIAID NIH HHS/United States
- R01 AI079056/AI/NIAID NIH HHS/United States
- R01 AI108634/AI/NIAID NIH HHS/United States
- S10 OD018174/OD/NIH HHS/United States
- R00 CA184043/CA/NCI NIH HHS/United States
- R56 AI079056/AI/NIAID NIH HHS/United States
- F32 GM128377/GM/NIGMS NIH HHS/United States
- R01 CA232347/CA/NCI NIH HHS/United States
- F31 CA220801/CA/NCI NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

