Complement genes contribute sex-biased vulnerability in diverse disorders
- PMID: 32499649
- PMCID: PMC7319891
- DOI: 10.1038/s41586-020-2277-x
Complement genes contribute sex-biased vulnerability in diverse disorders
Abstract
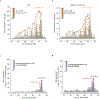
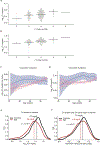
Many common illnesses, for reasons that have not been identified, differentially affect men and women. For instance, the autoimmune diseases systemic lupus erythematosus (SLE) and Sjögren's syndrome affect nine times more women than men1, whereas schizophrenia affects men with greater frequency and severity relative to women2. All three illnesses have their strongest common genetic associations in the major histocompatibility complex (MHC) locus, an association that in SLE and Sjögren's syndrome has long been thought to arise from alleles of the human leukocyte antigen (HLA) genes at that locus3-6. Here we show that variation of the complement component 4 (C4) genes C4A and C4B, which are also at the MHC locus and have been linked to increased risk for schizophrenia7, generates 7-fold variation in risk for SLE and 16-fold variation in risk for Sjögren's syndrome among individuals with common C4 genotypes, with C4A protecting more strongly than C4B in both illnesses. The same alleles that increase risk for schizophrenia greatly reduce risk for SLE and Sjögren's syndrome. In all three illnesses, C4 alleles act more strongly in men than in women: common combinations of C4A and C4B generated 14-fold variation in risk for SLE, 31-fold variation in risk for Sjögren's syndrome, and 1.7-fold variation in schizophrenia risk among men (versus 6-fold, 15-fold and 1.26-fold variation in risk among women, respectively). At a protein level, both C4 and its effector C3 were present at higher levels in cerebrospinal fluid and plasma8,9 in men than in women among adults aged between 20 and 50 years, corresponding to the ages of differential disease vulnerability. Sex differences in complement protein levels may help to explain the more potent effects of C4 alleles in men, women's greater risk of SLE and Sjögren's syndrome and men's greater vulnerability to schizophrenia. These results implicate the complement system as a source of sexual dimorphism in vulnerability to diverse illnesses.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




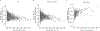
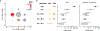


Comment in
-
Complement: the missing genetic link for SLE and pSS?Nat Rev Rheumatol. 2020 Aug;16(8):408. doi: 10.1038/s41584-020-0456-7. Nat Rev Rheumatol. 2020. PMID: 32561874 No abstract available.
-
Let's talk about sex.Sci Immunol. 2020 Jul 3;5(49):eabd4828. doi: 10.1126/sciimmunol.abd4828. Sci Immunol. 2020. PMID: 32620563
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH104964/MH/NIMH NIH HHS/United States
- U01 MH105653/MH/NIMH NIH HHS/United States
- MR/P005748/1/MRC_/Medical Research Council/United Kingdom
- R56 HG006855/HG/NHGRI NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- R01 MH123451/MH/NIMH NIH HHS/United States
- MR/L002604/1/MRC_/Medical Research Council/United Kingdom
- R01 MH085548/MH/NIMH NIH HHS/United States
- R01 HG006855/HG/NHGRI NIH HHS/United States
- P50 MH112491/MH/NIMH NIH HHS/United States
- G0901310/MRC_/Medical Research Council/United Kingdom
- U01 MH105641/MH/NIMH NIH HHS/United States
- MR/T04604X/1/MRC_/Medical Research Council/United Kingdom
- G0801418/MRC_/Medical Research Council/United Kingdom
- MR/L023784/2/MRC_/Medical Research Council/United Kingdom
- MR/L010305/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

