GC-AG Introns Features in Long Non-coding and Protein-Coding Genes Suggest Their Role in Gene Expression Regulation
- PMID: 32499820
- PMCID: PMC7242645
- DOI: 10.3389/fgene.2020.00488
GC-AG Introns Features in Long Non-coding and Protein-Coding Genes Suggest Their Role in Gene Expression Regulation
Abstract
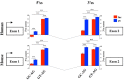
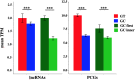
Long non-coding RNAs (lncRNAs) are recognized as an important class of regulatory molecules involved in a variety of biological functions. However, the regulatory mechanisms of long non-coding genes expression are still poorly understood. The characterization of the genomic features of lncRNAs is crucial to get insight into their function. In this study, we exploited recent annotations by GENCODE to characterize the genomic and splicing features of long non-coding genes in comparison with protein-coding ones, both in human and mouse. Our analysis highlighted differences between the two classes of genes in terms of their gene architecture. Significant differences in the splice sites usage were observed between long non-coding and protein-coding genes (PCG). While the frequency of non-canonical GC-AG splice junctions represents about 0.8% of total splice sites in PCGs, we identified a significant enrichment of the GC-AG splice sites in long non-coding genes, both in human (3.0%) and mouse (1.9%). In addition, we found a positional bias of GC-AG splice sites being enriched in the first intron in both classes of genes. Moreover, a significant shorter length and weaker donor and acceptor sites were found comparing GC-AG introns to GT-AG introns. Genes containing at least one GC-AG intron were found conserved in many species, more prone to alternative splicing and a functional analysis pointed toward their enrichment in specific biological processes such as DNA repair. Our study shows for the first time that GC-AG introns are mainly associated with lncRNAs and are preferentially located in the first intron. Additionally, we discovered their regulatory potential indicating the existence of a new mechanism of non-coding and PCGs expression regulation.
Keywords: GC-AG introns; alternative splicing; first intron; long non-coding RNAs; splice junctions.
Copyright © 2020 Abou Alezz, Celli, Belotti, Lisa and Bione.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

