Repurposing Modular Polyketide Synthases and Non-ribosomal Peptide Synthetases for Novel Chemical Biosynthesis
- PMID: 32500080
- PMCID: PMC7242659
- DOI: 10.3389/fmolb.2020.00087
Repurposing Modular Polyketide Synthases and Non-ribosomal Peptide Synthetases for Novel Chemical Biosynthesis
Abstract
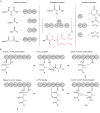
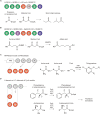
In nature, various enzymes govern diverse biochemical reactions through their specific three-dimensional structures, which have been harnessed to produce many useful bioactive compounds including clinical agents and commodity chemicals. Polyketide synthases (PKSs) and non-ribosomal peptide synthetases (NRPSs) are particularly unique multifunctional enzymes that display modular organization. Individual modules incorporate their own specific substrates and collaborate to assemble complex polyketides or non-ribosomal polypeptides in a linear fashion. Due to the modular properties of PKSs and NRPSs, they have been attractive rational engineering targets for novel chemical production through the predictable modification of each moiety of the complex chemical through engineering of the cognate module. Thus, individual reactions of each module could be separated as a retro-biosynthetic biopart and repurposed to new biosynthetic pathways for the production of biofuels or commodity chemicals. Despite these potentials, repurposing attempts have often failed owing to impaired catalytic activity or the production of unintended products due to incompatible protein-protein interactions between the modules and structural perturbation of the enzyme. Recent advances in the structural, computational, and synthetic tools provide more opportunities for successful repurposing. In this review, we focused on the representative strategies and examples for the repurposing of modular PKSs and NRPSs, along with their advantages and current limitations. Thereafter, synthetic biology tools and perspectives were suggested for potential further advancement, including the rational and large-scale high-throughput approaches. Ultimately, the potential diverse reactions from modular PKSs and NRPSs would be leveraged to expand the reservoir of useful chemicals.
Keywords: domain; module; non-ribosomal peptide synthetase; polyketide synthase; repurposing.
Copyright © 2020 Hwang, Lee, Cho, Palsson and Cho.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources

