SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts
- PMID: 32500504
- PMCID: PMC7271831
- DOI: 10.1007/s12250-020-00242-1
SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts
Abstract
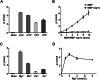
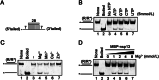
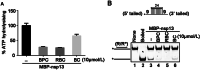
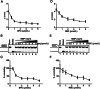
The ongoing outbreak of Coronavirus Disease 2019 (COVID-19) has become a global public health emergency. SARS-coronavirus-2 (SARS-CoV-2), the causative pathogen of COVID-19, is a positive-sense single-stranded RNA virus belonging to the family Coronaviridae. For RNA viruses, virus-encoded RNA helicases have long been recognized to play pivotal roles during viral life cycles by facilitating the correct folding and replication of viral RNAs. Here, our studies show that SARS-CoV-2-encoded nonstructural protein 13 (nsp13) possesses the nucleoside triphosphate hydrolase (NTPase) and RNA helicase activities that can hydrolyze all types of NTPs and unwind RNA helices dependently of the presence of NTP, and further characterize the biochemical characteristics of these two enzymatic activities associated with SARS-CoV-2 nsp13. Moreover, we found that some bismuth salts could effectively inhibit both the NTPase and RNA helicase activities of SARS-CoV-2 nsp13 in a dose-dependent manner. Thus, our findings demonstrate the NTPase and helicase activities of SARS-CoV-2 nsp13, which may play an important role in SARS-CoV-2 replication and serve as a target for antivirals.
Keywords: Antiviral target; Helicase; NTPase; Nsp13; SARS-coronavirus-2 (SARS-CoV-2).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


Similar articles
-
SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists.Emerg Microbes Infect. 2020 Dec;9(1):1418-1428. doi: 10.1080/22221751.2020.1780953. Emerg Microbes Infect. 2020. PMID: 32529952 Free PMC article.
-
Evaluation of the potency of FDA-approved drugs on wild type and mutant SARS-CoV-2 helicase (Nsp13).Int J Biol Macromol. 2020 Nov 15;163:1687-1696. doi: 10.1016/j.ijbiomac.2020.09.138. Epub 2020 Sep 24. Int J Biol Macromol. 2020. PMID: 32980406 Free PMC article.
-
The stalk domain of SARS-CoV-2 NSP13 is essential for its helicase activity.Biochem Biophys Res Commun. 2022 Apr 23;601:129-136. doi: 10.1016/j.bbrc.2022.02.068. Epub 2022 Feb 23. Biochem Biophys Res Commun. 2022. PMID: 35245742 Free PMC article.
-
The Coronavirus helicase in replication.Virus Res. 2024 Aug;346:199401. doi: 10.1016/j.virusres.2024.199401. Epub 2024 May 31. Virus Res. 2024. PMID: 38796132 Free PMC article. Review.
-
A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping.Cells. 2020 May 20;9(5):1267. doi: 10.3390/cells9051267. Cells. 2020. PMID: 32443810 Free PMC article. Review.
Cited by
-
A Comprehensive View on the Protein Functions of Porcine Epidemic Diarrhea Virus.Genes (Basel). 2024 Jan 26;15(2):165. doi: 10.3390/genes15020165. Genes (Basel). 2024. PMID: 38397155 Free PMC article. Review.
-
Understanding Individual SARS-CoV-2 Proteins for Targeted Drug Development against COVID-19.Mol Cell Biol. 2021 Aug 24;41(9):e0018521. doi: 10.1128/MCB.00185-21. Epub 2021 Aug 24. Mol Cell Biol. 2021. PMID: 34124934 Free PMC article. Review.
-
COVID-19: molecular targets, drug repurposing and new avenues for drug discovery.Mem Inst Oswaldo Cruz. 2020 Oct 2;115:e200254. doi: 10.1590/0074-02760200254. eCollection 2020. Mem Inst Oswaldo Cruz. 2020. PMID: 33027420 Free PMC article.
-
Molecular mechanisms of the novel coronavirus SARS-CoV-2 and potential anti-COVID19 pharmacological targets since the outbreak of the pandemic.Food Chem Toxicol. 2020 Dec;146:111805. doi: 10.1016/j.fct.2020.111805. Epub 2020 Oct 8. Food Chem Toxicol. 2020. PMID: 33038452 Free PMC article. Review.
-
SARS-CoV-2 Nsp14 mediates the effects of viral infection on the host cell transcriptome.Elife. 2022 Mar 16;11:e71945. doi: 10.7554/eLife.71945. Elife. 2022. PMID: 35293857 Free PMC article.
References
-
- Adedeji AO, Singh K, Calcaterra NE, DeDiego ML, Enjuanes L, Weiss S, Sarafianos SG. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob Agents Chemother. 2012;56:4718–4728. doi: 10.1128/AAC.00957-12. - DOI - PMC - PubMed
-
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

