miR-623 suppresses cell proliferation, migration and invasion through direct inhibition of XRCC5 in breast cancer
- PMID: 32501811
- PMCID: PMC7346019
- DOI: 10.18632/aging.103182
miR-623 suppresses cell proliferation, migration and invasion through direct inhibition of XRCC5 in breast cancer
Abstract
Background/aims: MicroRNAs (miRNAs) are short, non-coding RNA molecules that control gene expression trough negative translational regulation. MiR-623 is a tumor suppressor, and it's function and mechanism in breast cancer has not been reported.
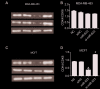
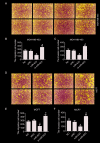
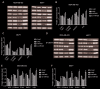
Results: Exogenous overexpression of miR-623 suppressed cell proliferation, migration and invasion, meanwhile, but promoted cell apoptosis. MiR-623 knockdown displayed opposite results. Overexpression of miR-623 resulted in the downregulation of CDK4/6 as well as the inhibition of the phosphatidylinositol-3-kinase (PI3K)/Akt and Wnt/β-Catenin signaling pathways. MiR-623 knockdown displayed opposite results. Results of the reporter assay revealed that the luciferase activity was decreased in XRCC5-wt cells, suggesting that miR-623 could directly combine with 3' UTR of XRCC5. MiR-623 significantly suppressed XRCC5 expression, which is critical for miR-623-induced proliferation and migration block in breast cancer cells.
Conclusion: miR-623 suppressed cell proliferation, migration and invasion through downregulation of cyclin dependent kinases and inhibition of the phosphatidylinositol-3-kinase (PI3K)/Akt and Wnt/β-Catenin pathways by targeting XRCC5.
Methods: miR-623 was either overexpressed in breast cancer cell lines through exogenous transfection or knocked down by specific siRNA. Cell proliferation, migration and invasion were examined using CCK-8, colony formation and transwell assay. The direct target of miR-623 was verified using luciferase reporter gene assay.
Keywords: XRCC5; breast cancer; microRNA; migration; proliferation.
Conflict of interest statement
Figures



Similar articles
-
In vivo and in vitro effects of microRNA-27a on proliferation, migration and invasion of breast cancer cells through targeting of SFRP1 gene via Wnt/β-catenin signaling pathway.Oncotarget. 2017 Feb 28;8(9):15507-15519. doi: 10.18632/oncotarget.14662. Oncotarget. 2017. PMID: 28099945 Free PMC article.
-
Overexpression of miR-623 suppresses progression of hepatocellular carcinoma via regulating the PI3K/Akt signaling pathway by targeting XRCC5.J Cell Biochem. 2020 Jan;121(1):213-223. doi: 10.1002/jcb.29117. Epub 2019 Jun 12. J Cell Biochem. 2020. PMID: 31190447
-
MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway.Tumour Biol. 2016 Jul;37(7):8993-9000. doi: 10.1007/s13277-015-4513-9. Epub 2016 Jan 12. Tumour Biol. 2016. PMID: 26758430
-
Dysregulation of miR-411 in cancer: Causative factor for pathogenesis, diagnosis and prognosis.Biomed Pharmacother. 2022 May;149:112896. doi: 10.1016/j.biopha.2022.112896. Epub 2022 Mar 29. Biomed Pharmacother. 2022. PMID: 35358797 Review.
-
Functional mechanism and clinical implications of miR-141 in human cancers.Cell Signal. 2022 Jul;95:110354. doi: 10.1016/j.cellsig.2022.110354. Epub 2022 May 10. Cell Signal. 2022. PMID: 35550172 Review.
Cited by
-
Construction of a miRNA-mRNA Network Related to Exosomes in Colon Cancer.Dis Markers. 2022 Jul 5;2022:2192001. doi: 10.1155/2022/2192001. eCollection 2022. Dis Markers. 2022. PMID: 35845138 Free PMC article.
-
HIF-1α-Mediated miR-623 Regulates Apoptosis and Inflammatory Responses of Nucleus Pulposus Induced by Oxidative Stress via Targeting TXNIP.Oxid Med Cell Longev. 2021 Aug 3;2021:6389568. doi: 10.1155/2021/6389568. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34394829 Free PMC article.
-
DNA Damage Response in Cancer Therapy and Resistance: Challenges and Opportunities.Int J Mol Sci. 2022 Nov 24;23(23):14672. doi: 10.3390/ijms232314672. Int J Mol Sci. 2022. PMID: 36499000 Free PMC article. Review.
-
A review on the role of cyclin dependent kinases in cancers.Cancer Cell Int. 2022 Oct 20;22(1):325. doi: 10.1186/s12935-022-02747-z. Cancer Cell Int. 2022. PMID: 36266723 Free PMC article. Review.
-
Grapefruit Juice Flavanones Modulate the Expression of Genes Regulating Inflammation, Cell Interactions and Vascular Function in Peripheral Blood Mononuclear Cells of Postmenopausal Women.Front Nutr. 2022 May 26;9:907595. doi: 10.3389/fnut.2022.907595. eCollection 2022. Front Nutr. 2022. PMID: 35694160 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

