Homozygous Mutations in BTG4 Cause Zygotic Cleavage Failure and Female Infertility
- PMID: 32502391
- PMCID: PMC7332666
- DOI: 10.1016/j.ajhg.2020.05.010
Homozygous Mutations in BTG4 Cause Zygotic Cleavage Failure and Female Infertility
Abstract
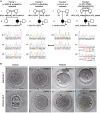
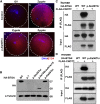
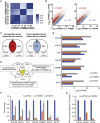
Zygotic cleavage failure (ZCF) is a unique early embryonic phenotype resulting in female infertility and recurrent failure of in vitro fertilization (IVF) and/or intracytoplasmic sperm injection (ICSI). With this phenotype, morphologically normal oocytes can be retrieved and successfully fertilized, but they fail to undergo cleavage. Until now, whether this phenotype has a Mendelian inheritance pattern and which underlying genetic factors play a role in its development remained to be elucidated. B cell translocation gene 4 (BTG4) is a key adaptor of the CCR4-NOT deadenylase complex, which is involved in maternal mRNA decay in mice, but no human diseases caused by mutations in BTG4 have previously been reported. Here, we identified four homozygous mutations in BTG4 (GenBank: NM_017589.4) that are responsible for the phenotype of ZCF, and we found they followed a recessive inheritance pattern. Three of them-c.73C>T (p.Gln25Ter), c.1A>G (p.?), and c.475_478del (p.Ile159LeufsTer15)-resulted in complete loss of full-length BTG4 protein. For c.166G>A (p.Ala56Thr), although the protein level and distribution of mutant BTG4 was not altered in zygotes from affected individuals or in HeLa cells, the interaction between BTG4 and CNOT7 was abolished. In vivo studies further demonstrated that the process of maternal mRNA decay was disrupted in the zygotes of the affected individuals, which provides a mechanistic explanation for the phenotype of ZCF. Thus, we provide evidence that ZCF is a Mendelian phenotype resulting from mutations in BTG4. These findings contribute to our understanding of the role of BTG4 in human early embryonic development and provide a genetic marker for female infertility.
Keywords: BTG4; female infertility; mutation; zygotic cleavage failure.
Copyright © 2020 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hart R.J. Physiological aspects of female fertility: role of the environment, modern lifestyle, and genetics. Physiol. Rev. 2016;96:873–909. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

