Rad54 Drives ATP Hydrolysis-Dependent DNA Sequence Alignment during Homologous Recombination
- PMID: 32502392
- PMCID: PMC7418177
- DOI: 10.1016/j.cell.2020.04.056
Rad54 Drives ATP Hydrolysis-Dependent DNA Sequence Alignment during Homologous Recombination
Abstract
Homologous recombination (HR) helps maintain genome integrity, and HR defects give rise to disease, especially cancer. During HR, damaged DNA must be aligned with an undamaged template through a process referred to as the homology search. Despite decades of study, key aspects of this search remain undefined. Here, we use single-molecule imaging to demonstrate that Rad54, a conserved Snf2-like protein found in all eukaryotes, switches the search from the diffusion-based pathways characteristic of the basal HR machinery to an active process in which DNA sequences are aligned via an ATP-dependent molecular motor-driven mechanism. We further demonstrate that Rad54 disrupts the donor template strands, enabling the search to take place within a migrating DNA bubble-like structure that is bound by replication protein A (RPA). Our results reveal that Rad54, working together with RPA, fundamentally alters how DNA sequences are aligned during HR.
Keywords: DNA curtain; DNA double-strand break; Rad51; Rad54; homologous recombination; homology search; motor protein; replication protein A; single molecule.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

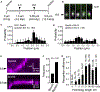
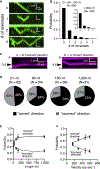
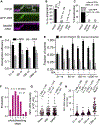
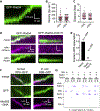


References
-
- Adzuma K (1998). No sliding during homology search by RecA protein. J. Biol. Chem 273, 31565–31573. - PubMed
-
- Alexeev A, Mazin A, and Kowalczykowski SC (2003). Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol 10, 182–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

