Pathogenic Allodiploid Hybrids of Aspergillus Fungi
- PMID: 32502407
- PMCID: PMC7343619
- DOI: 10.1016/j.cub.2020.04.071
Pathogenic Allodiploid Hybrids of Aspergillus Fungi
Abstract
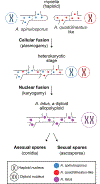
Interspecific hybridization substantially alters genotypes and phenotypes and can give rise to new lineages. Hybrid isolates that differ from their parental species in infection-relevant traits have been observed in several human-pathogenic yeasts and plant-pathogenic filamentous fungi but have yet to be found in human-pathogenic filamentous fungi. We discovered 6 clinical isolates from patients with aspergillosis originally identified as Aspergillus nidulans (section Nidulantes) that are actually allodiploid hybrids formed by the fusion of Aspergillus spinulosporus with an unknown close relative of Aspergillus quadrilineatus, both in section Nidulantes. Evolutionary genomic analyses revealed that these isolates belong to Aspergillus latus, an allodiploid hybrid species. Characterization of diverse infection-relevant traits further showed that A. latus hybrid isolates are genomically and phenotypically heterogeneous but also differ from A. nidulans, A. spinulosporus, and A. quadrilineatus. These results suggest that allodiploid hybridization contributes to the genomic and phenotypic diversity of filamentous fungal pathogens of humans.
Keywords: Eurotiomycetes; allopolyploidy; ascomycota; aspergillosis; cryptic species; fungal pathogen evolution; hybridization; nonvertical evolution; pathogenicity; virulence.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
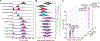


References
-
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J,Brelsford A, Buerkle CA, Buggs R, et al. (2013). Hybridization and speciation. J. Evol. Biol. 26, 229–246. - PubMed
-
- Rieseberg LH (2003). Major Ecological Transitions in Wild Sunflowers Facilitated by Hybridization. Science (80-. ). 301, 1211–1216. - PubMed
-
- Mable BK, Alexandrou MA, and Taylor MI (2011). Genome duplication in amphibians and fish: an extended synthesis. J. Zool. 284, 151–182.
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

