Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface
- PMID: 32502616
- PMCID: PMC7267807
- DOI: 10.1016/j.jtos.2020.05.013
Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface
Abstract
Purpose: The high infection rate of SARS-CoV-2 necessitates the need for multiple studies identifying the molecular mechanisms that facilitate the viral entry and propagation. Currently the potential extra-respiratory transmission routes of SARS-CoV-2 remain unclear.
Methods: Using single-cell RNA Seq and ATAC-Seq datasets and immunohistochemical analysis, we investigated SARS-CoV-2 tropism in the embryonic, fetal and adult human ocular surface.
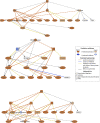
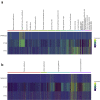
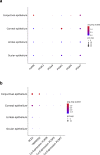
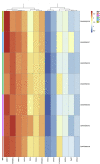
Results: The co-expression of ACE2 receptor and entry protease TMPRSS2 was detected in the human adult conjunctival, limbal and corneal epithelium, but not in the embryonic and fetal ocular surface up to 21 post conception weeks. These expression patterns were corroborated by the single cell ATAC-Seq data, which revealed a permissive chromatin in ACE2 and TMPRSS2 loci in the adult conjunctival, limbal and corneal epithelium. Co-expression of ACE2 and TMPRSS2 was strongly detected in the superficial limbal, corneal and conjunctival epithelium, implicating these as target entry cells for SARS-CoV-2 in the ocular surface. Strikingly, we also identified the key pro-inflammatory signals TNF, NFKβ and IFNG as upstream regulators of the transcriptional profile of ACE2+TMPRSS2+ cells in the superficial conjunctival epithelium, suggesting that SARS-CoV-2 may utilise inflammatory driven upregulation of ACE2 and TMPRSS2 expression to enhance infection in ocular surface.
Conclusions: Together our data indicate that the human ocular surface epithelium provides an additional entry portal for SARS-CoV-2, which may exploit inflammatory driven upregulation of ACE2 and TMPRSS2 entry factors to enhance infection.
Keywords: Conjunctiva; Cornea; ace2; sars-cov-2; tmprss2.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
None.
Figures







Comment in
-
SARS-CoV-2 receptor ACE2 is expressed in human conjunctival tissue, especially in diseased conjunctival tissue.Ocul Surf. 2021 Jan;19:249-251. doi: 10.1016/j.jtos.2020.09.010. Epub 2020 Sep 28. Ocul Surf. 2021. PMID: 32991984 Free PMC article. No abstract available.
References
-
- Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technica...
-
- COVID-19 map - Johns Hopkins coronavirus Resource center. https://coronavirus.jhu.edu/map.html
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

