Differential Effects of Human Adenovirus E1A Protein Isoforms on Aerobic Glycolysis in A549 Human Lung Epithelial Cells
- PMID: 32503156
- PMCID: PMC7354625
- DOI: 10.3390/v12060610
Differential Effects of Human Adenovirus E1A Protein Isoforms on Aerobic Glycolysis in A549 Human Lung Epithelial Cells
Abstract
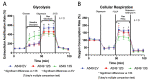
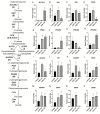
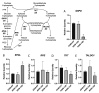
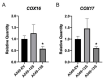
Viruses alter a multitude of host-cell processes to create a more optimal environment for viral replication. This includes altering metabolism to provide adequate substrates and energy required for replication. Typically, viral infections induce a metabolic phenotype resembling the Warburg effect, with an upregulation of glycolysis and a concurrent decrease in cellular respiration. Human adenovirus (HAdV) has been observed to induce the Warburg effect, which can be partially attributed to the adenovirus protein early region 4, open reading frame 1 (E4orf1). E4orf1 regulates a multitude of host-cell processes to benefit viral replication and can influence cellular metabolism through the transcription factor avian myelocytomatosis viral oncogene homolog (MYC). However, E4orf1 does not explain the full extent of Warburg-like HAdV metabolic reprogramming, especially the accompanying decrease in cellular respiration. The HAdV protein early region 1A (E1A) also modulates the function of the infected cell to promote viral replication. E1A can interact with a wide variety of host-cell proteins, some of which have been shown to interact with metabolic enzymes independently of an interaction with E1A. To determine if the HAdV E1A proteins are responsible for reprogramming cell metabolism, we measured the extracellular acidification rate and oxygen consumption rate of A549 human lung epithelial cells with constitutive endogenous expression of either of the two major E1A isoforms. This was followed by the characterization of transcript levels for genes involved in glycolysis and cellular respiration, and related metabolic pathways. Cells expressing the 13S encoded E1A isoform had drastically increased baseline glycolysis and lower maximal cellular respiration than cells expressing the 12S encoded E1A isoform. Cells expressing the 13S encoded E1A isoform exhibited upregulated expression of glycolysis genes and downregulated expression of cellular respiration genes. However, tricarboxylic acid cycle genes were upregulated, resembling anaplerotic metabolism employed by certain cancers. Upregulation of glycolysis and tricarboxylic acid cycle genes was also apparent in IMR-90 human primary lung fibroblast cells infected with a HAdV-5 mutant virus that expressed the 13S, but not the 12S encoded E1A isoform. In conclusion, it appears that the two major isoforms of E1A differentially influence cellular glycolysis and oxidative phosphorylation and this is at least partially due to the altered regulation of mRNA expression for the genes in these pathways.
Keywords: 12S; 13S; E1A; Warburg effect; cellular respiration; glycolysis; human adenovirus; oxidative phosphorylation; pentose phosphate pathway; tricarboxylic acid cycle.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Warburg O. The metabolism of carcinoma cells. J. Cancer Res. 1925;9:148–163. doi: 10.1158/jcr.1925.148. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

