Archaeal DNA Replication
- PMID: 32503372
- PMCID: PMC7712474
- DOI: 10.1146/annurev-micro-020518-115443
Archaeal DNA Replication
Abstract
It is now well recognized that the information processing machineries of archaea are far more closely related to those of eukaryotes than to those of their prokaryotic cousins, the bacteria. Extensive studies have been performed on the structure and function of the archaeal DNA replication origins, the proteins that define them, and the macromolecular assemblies that drive DNA unwinding and nascent strand synthesis. The results from various archaeal organisms across the archaeal domain of life show surprising levels of diversity at many levels-ranging from cell cycle organization to chromosome ploidy to replication mode and nature of the replicative polymerases. In the following, we describe recent advances in the field, highlighting conserved features and lineage-specific innovations.
Keywords: CMG; DNA polymerase; DNA primase; Sulfolobus; archaea; helicase.
Figures
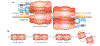
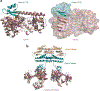
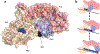
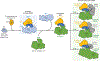
References
-
- Bell SD. 2017. Initiation of DNA replication in the archaea. Adv. Exp. Med. Biol 1042:99–115 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

