Identification of novel alternative splicing isoform biomarkers and their association with overall survival in colorectal cancer
- PMID: 32503434
- PMCID: PMC7275609
- DOI: 10.1186/s12876-020-01288-x
Identification of novel alternative splicing isoform biomarkers and their association with overall survival in colorectal cancer
Abstract
Background: Alternative splicing (AS) is an important mechanism of regulating eukaryotic gene expression. Understanding the most common AS events in colorectal cancer (CRC) will help developing diagnostic, prognostic or therapeutic tools in CRC.
Methods: Publicly available RNA-seq data of 28 pairs of CRC and normal tissues and 18 pairs of metastatic and normal tissues were used to identify AS events using PSI and DEXSeq methods.
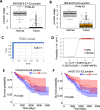
Result: The highly significant splicing events were used to search a database of The Cancer Genome Atlas (TCGA). We identified AS events in 9 genes in CRC (more inclusion of CLK1-E4, COL6A3-E6, CD44v8-10, alternative first exon regulation of ARHGEF9, CHEK1, HKDC1 and HNF4A) or metastasis (decrease of SERPINA1-E1a, CALD-E5b, E6). Except for CHEK1, all other 8 splicing events were confirmed by TCGA data with 382 CRC tumors and 51 normal controls. The combination of three splicing events was used to build a logistic regression model that can predict sample type (CRC or normal) with near perfect performance (AUC = 1). Two splicing events (COL6A3 and HKDC1) were found to be significantly associated with patient overall survival. The AS features of the 9 genes are highly consistent with previous reports and/or relevant to cancer biology.
Conclusions: The significant association of higher expression of the COL6A3 E5-E6 junction and HKDC1 E1-E2 with better overall survival was firstly reported. This study might be of significant value in the future biomarker, prognosis marker and therapeutics development of CRC.
Keywords: Alternative splicing (AS); Colorectal cancer (CRC); Metastasis; RNA-seq; TCGA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

