Multi-trait analysis of rare-variant association summary statistics using MTAR
- PMID: 32503972
- PMCID: PMC7275056
- DOI: 10.1038/s41467-020-16591-0
Multi-trait analysis of rare-variant association summary statistics using MTAR
Abstract
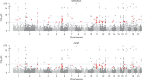
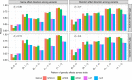
Integrating association evidence across multiple traits can improve the power of gene discovery and reveal pleiotropy. Most multi-trait analysis methods focus on individual common variants in genome-wide association studies. Here, we introduce multi-trait analysis of rare-variant associations (MTAR), a framework for joint analysis of association summary statistics between multiple rare variants and different traits. MTAR achieves substantial power gain by leveraging the genome-wide genetic correlation measure to inform the degree of gene-level effect heterogeneity across traits. We apply MTAR to rare-variant summary statistics for three lipid traits in the Global Lipids Genetics Consortium. 99 genome-wide significant genes were identified in the single-trait-based tests, and MTAR increases this to 139. Among the 11 novel lipid-associated genes discovered by MTAR, 7 are replicated in an independent UK Biobank GWAS analysis. Our study demonstrates that MTAR is substantially more powerful than single-trait-based tests and highlights the value of MTAR for novel gene discovery.
Conflict of interest statement
J.S., H.Z., A.C., and D.V.M. are employees at Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

