A synthesis of bacterial and archaeal phenotypic trait data
- PMID: 32503990
- PMCID: PMC7275036
- DOI: 10.1038/s41597-020-0497-4
A synthesis of bacterial and archaeal phenotypic trait data
Abstract
A synthesis of phenotypic and quantitative genomic traits is provided for bacteria and archaea, in the form of a scripted, reproducible workflow that standardizes and merges 26 sources. The resulting unified dataset covers 14 phenotypic traits, 5 quantitative genomic traits, and 4 environmental characteristics for approximately 170,000 strain-level and 15,000 species-aggregated records. It spans all habitats including soils, marine and fresh waters and sediments, host-associated and thermal. Trait data can find use in clarifying major dimensions of ecological strategy variation across species. They can also be used in conjunction with species and abundance sampling to characterize trait mixtures in communities and responses of traits along environmental gradients.
Conflict of interest statement
The authors declare no competing interests.
Figures
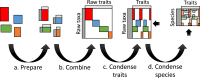

References
-
- Litchman E, Klausmeier CA. Trait-Based Community Ecology of Phytoplankton. Annu. Rev. Ecol. Evol. S. 2008;39:615–639. doi: 10.1146/annurev.ecolsys.39.110707.173549. - DOI
-
- Litchman E, et al. Global biogeochemical impacts of phytoplankton: a trait-based perspective. J. Ecol. 2015;103:1384–1396. doi: 10.1111/1365-2745.12438. - DOI
-
- Martiny, J. B. H., Jones, S. E., Lennon, J. T. & Martiny, A. C. Microbiomes in light of traits: A phylogenetic perspective. Science 350, aac9323 (2015). - PubMed

