Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair
- PMID: 32504093
- PMCID: PMC7399532
- DOI: 10.1093/hmg/ddaa106
Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair
Abstract
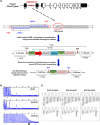
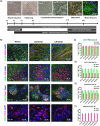
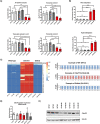
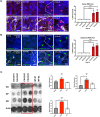
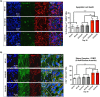
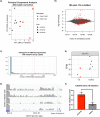
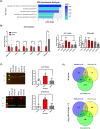
The G4C2 hexanucleotide repeat expansion (HRE) in C9orf72 is the commonest cause of familial amyotrophic lateral sclerosis (ALS). A number of different methods have been used to generate isogenic control lines using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 and non-homologous end-joining by deleting the repeat region, with the risk of creating indels and genomic instability. In this study, we demonstrate complete correction of an induced pluripotent stem cell (iPSC) line derived from a C9orf72-HRE positive ALS/frontotemporal dementia patient using CRISPR/Cas9 genome editing and homology-directed repair (HDR), resulting in replacement of the excised region with a donor template carrying the wild-type repeat size to maintain the genetic architecture of the locus. The isogenic correction of the C9orf72 HRE restored normal gene expression and methylation at the C9orf72 locus, reduced intron retention in the edited lines and abolished pathological phenotypes associated with the C9orf72 HRE expansion in iPSC-derived motor neurons (iPSMNs). RNA sequencing of the mutant line identified 2220 differentially expressed genes compared with its isogenic control. Enrichment analysis demonstrated an over-representation of ALS relevant pathways, including calcium ion dependent exocytosis, synaptic transport and the Kyoto Encyclopedia of Genes and Genomes ALS pathway, as well as new targets of potential relevance to ALS pathophysiology. Complete correction of the C9orf72 HRE in iPSMNs by CRISPR/Cas9-mediated HDR provides an ideal model to study the earliest effects of the hexanucleotide expansion on cellular homeostasis and the key pathways implicated in ALS pathophysiology.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Chio A., Mora G., Leone M., Mazzini L., Cocito D., Giordana M.T., Bottacchi E., Mutani R. and Piemonte and Valle d’Aosta Register for ALS (2002) Early symptom progression rate is related to ALS outcome: a prospective population-based study. Neurology, 59, 99–103. - PubMed
-
- Majounie E., Renton A.E., Mok K., Dopper E.G.P.P., Waite A., Rollinson S., Chiò A., Restagno G., Nicolaou N., Simon-Sanchez J. et al. (2012) Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol., 11, 323–330. - PMC - PubMed
-
- van Blitterswijk M., DeJesus-Hernandez M., Niemantsverdriet E., Murray M.E., Heckman M.G., Diehl N.N., Brown P.H., Baker M.C., Finch N.A., Bauer P.O. et al. (2013) Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol., 12, 978–988. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- TALBOT/JULY13/820-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- CL-2015-26-001/DH_/Department of Health/United Kingdom
- TURNER/OCT18/989-797/MNDA_/Motor Neurone Disease Association/United Kingdom
- MR/L002167/1/MRC_/Medical Research Council/United Kingdom
- MR/L023784/1/MRC_/Medical Research Council/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- MC_EX_MR/N50192X/1/MRC_/Medical Research Council/United Kingdom
- MR/M024962/1/MRC_/Medical Research Council/United Kingdom
- TALBOT-MUTIHAC/APR15/832-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- MC_PC_15065/MRC_/Medical Research Council/United Kingdom
- TALBOT/OCT16/889-792/MNDA_/Motor Neurone Disease Association/United Kingdom
- WTISSF121302/WT_/Wellcome Trust/United Kingdom
- J-0901/PUK_/Parkinson's UK/United Kingdom
- SCABER/JULY13/945-795/MNDA_/Motor Neurone Disease Association/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/P007023/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

