Phosphorylation of mitochondrial transcription factor B2 controls mitochondrial DNA binding and transcription
- PMID: 32505352
- PMCID: PMC9161741
- DOI: 10.1016/j.bbrc.2020.05.141
Phosphorylation of mitochondrial transcription factor B2 controls mitochondrial DNA binding and transcription
Abstract
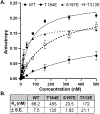
Mammalian cells contain genetic information in two compartments, the nucleus and the mitochondria. Mitochondrial gene expression must be coordinated with nuclear gene expression to respond to cellular energetic needs. To gain insight into the coordination between the nucleus and mitochondria, there is a need to understand the regulation of transcription of mitochondrial DNA (mtDNA). Reversible protein post-translational modifications of the mtDNA transcriptional machinery may be one way to control mtDNA transcription. Here we focus on a member of the mtDNA transcription initiation complex, mitochondrial transcription factor B2 (TFB2M). TFB2M melts mtDNA at the promoter to allow the RNA polymerase (POLRMT) to access the DNA template and initiate transcription. Three phosphorylation sites have been previously identified on TFB2M by mass spectrometry: threonine 184, serine 197, and threonine 313. Phosphomimetics were established at these positions. Proteins were purified and analyzed for their ability to bind mtDNA and initiate transcription in vitro. Our results indicate phosphorylation at threonine 184 and threonine 313 impairs promoter binding and prevents transcription. These findings provide a potential regulatory mechanism of mtDNA transcription and help clarify the importance of protein post-translational modifications in mitochondrial function.
Keywords: Mitochondrial DNA; Mitochondrial transcription factor B2 (TFB2M); Phosphorylation; Transcription regulation.
Copyright © 2020 Elsevier Inc. All rights reserved.
Figures

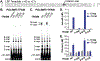

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

