Molecular analysis of several in-house rRT-PCR protocols for SARS-CoV-2 detection in the context of genetic variability of the virus in Colombia
- PMID: 32505692
- PMCID: PMC7272177
- DOI: 10.1016/j.meegid.2020.104390
Molecular analysis of several in-house rRT-PCR protocols for SARS-CoV-2 detection in the context of genetic variability of the virus in Colombia
Abstract
The COVID-19 pandemic caused by SARS-CoV-2 is a public health problem unprecedented in the recent history of humanity. Different in-house real-time RT-PCR (rRT-PCR) methods for SARS-CoV-2 diagnosis and the appearance of genomes with mutations in primer regions have been reported. Hence, whole-genome data from locally-circulating SARS-CoV-2 strains contribute to the knowledge of its global variability and the development and fine tuning of diagnostic protocols. To describe the genetic variability of Colombian SARS-CoV-2 genomes in hybridization regions of oligonucleotides of the main in-house methods for SARS-CoV-2 detection, RNA samples with confirmed SARS-CoV-2 molecular diagnosis were processed through next-generation sequencing. Primers/probes sequences from 13 target regions for SARS-CoV-2 detection suggested by 7 institutions and consolidated by WHO during the early stage of the pandemic were aligned with Muscle tool to assess the genetic variability potentially affecting their performance. Finally, the corresponding codon positions at the 3' end of each primer, the open reading frame inspection was identified for each gene/protein product. Complete SARS-CoV-2 genomes were obtained from 30 COVID-19 cases, representative of the current epidemiology in the country. Mismatches between at least one Colombian sequence and five oligonucleotides targeting the RdRP and N genes were observed. The 3' end of 4 primers aligned to the third codon position, showed high risk of nucleotide substitution and potential mismatches at this critical position. Genetic variability was detected in Colombian SARS-CoV-2 sequences in some of the primer/probe regions for in-house rRT-PCR diagnostic tests available at WHO COVID-19 technical guidelines; its impact on the performance and rates of false-negative results should be experimentally evaluated. The genomic surveillance of SARS-CoV-2 is highly recommended for the early identification of mutations in critical regions and to issue recommendations on specific diagnostic tests to ensure the coverage of locally-circulating genetic variants.
Keywords: COVID-19 diagnostic testing; Genetic diversity; Next-generation sequencing; RT-PCR; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that there is no conflict of interest in the manuscript.
Figures






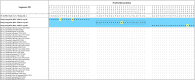

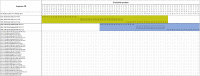


References
-
- BII/GIS . 2020. Analysis Update. (GISAID)
-
- brian-jgi . 2020. BBMap Short Read Aligner, and Other Bioinformatic Tools.
-
- Corman V.M., Rasche A., Baronti C., Aldabbagh S., Cadar D., Reusken C.B., Pas S.D., Goorhuis A., Schinkel J., Molenkamp R., Kummerer B.M., Bleicker T., Brunink S., Eschbach-Bludau M., Eis-Hubinger A.M., Koopmans M.P., Schmidt-Chanasit J., Grobusch M.P., de Lamballerie X., Drosten C., Drexler J.F. Assay optimization for molecular detection of Zika virus. Bull. World Health Organ. 2016;94:880–892. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

