Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays
- PMID: 32505777
- PMCID: PMC7263247
- DOI: 10.1016/j.jcv.2020.104480
Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays
Abstract
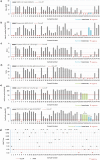
Serological SARS-CoV-2 assays are urgently needed for diagnosis, contact tracing and for epidemiological studies. So far, there is limited data on how recently commercially available, high-throughput immunoassays, using different recombinant SARS-CoV-2 antigens, perform with clinical samples. Focusing on IgG and total antibodies, we demonstrate the performance of four automated immunoassays (Abbott Architect™ i2000 (N protein-based)), Roche cobas™ e 411 analyzer (N protein-based, not differentiating between IgA, IgM or IgG antibodies), LIAISON®XL platform (S1 and S2 protein-based), VIRCLIA® automation system (S1 and N protein-based) in comparison to two ELISA assays (Euroimmun SARS-CoV-2 IgG (S1 protein-based) and Virotech SARS-CoV-2 IgG ELISA (N protein-based)) and an in-house developed plaque reduction neutralization test (PRNT). We tested follow up serum/plasma samples of individuals PCR-diagnosed with COVID-19. When calculating the overall sensitivity, in a time frame of 49 days after first PCR-positivity, the PRNT as gold standard, showed the highest sensitivity with 93.3% followed by the dual-target assay for the VIRCLIA® automation system with 89%. The overall sensitivity in the group of N protein-based assays ranged from 66.7 to 77.8% and in the S protein-based-assays from 71.1 to 75.6%. Five follow-up samples of three individuals were only detected in either an S and/or N protein-based assay, indicating an individual different immune response to SARS-CoV-2 and the influence of the used assay in the detection of IgG antibodies. This should be further analysed. The specificity of the examined assays was ≥ 97%. However, because of the low or unknown prevalence of SARS-CoV-2, the examined assays in this study are currently primarily eligible for epidemiological investigations, as they have limited information in individual testing.
Keywords: Antibody; Assay; Evaluation; IgG; PRNT; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest Sandra Ciesek received a speaker’s fee from Euroimmun. The other authors declare no conflicts of interest.
Figures
Comment in
-
Serologic aspects of COVID-19: Recommendations for use in the clinical setting.Travel Med Infect Dis. 2021 May-Jun;41:102046. doi: 10.1016/j.tmaid.2021.102046. Epub 2021 Mar 31. Travel Med Infect Dis. 2021. PMID: 33798744 Free PMC article. No abstract available.
References
-
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. - DOI - PMC - PubMed
-
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

