Healthy versus pathological learning transferability in shoulder muscle MRI segmentation using deep convolutional encoder-decoders
- PMID: 32505943
- PMCID: PMC9926537
- DOI: 10.1016/j.compmedimag.2020.101733
Healthy versus pathological learning transferability in shoulder muscle MRI segmentation using deep convolutional encoder-decoders
Abstract
Fully-automated segmentation of pathological shoulder muscles in patients with musculo-skeletal diseases is a challenging task due to the huge variability in muscle shape, size, location, texture and injury. A reliable automatic segmentation method from magnetic resonance images could greatly help clinicians to diagnose pathologies, plan therapeutic interventions and predict interventional outcomes while eliminating time consuming manual segmentation. The purpose of this work is three-fold. First, we investigate the feasibility of automatic pathological shoulder muscle segmentation using deep learning techniques, given a very limited amount of available annotated pediatric data. Second, we address the learning transferability from healthy to pathological data by comparing different learning schemes in terms of model generalizability. Third, extended versions of deep convolutional encoder-decoder architectures using encoders pre-trained on non-medical data are proposed to improve the segmentation accuracy. Methodological aspects are evaluated in a leave-one-out fashion on a dataset of 24 shoulder examinations from patients with unilateral obstetrical brachial plexus palsy and focus on 4 rotator cuff muscles (deltoid, infraspinatus, supraspinatus and subscapularis). The most accurate segmentation model is partially pre-trained on the large-scale ImageNet dataset and jointly exploits inter-patient healthy and pathological annotated data. Its performance reaches Dice scores of 82.4%, 82.0%, 71.0% and 82.8% for deltoid, infraspinatus, supraspinatus and subscapularis muscles. Absolute surface estimation errors are all below 83 mm2 except for supraspinatus with 134.6 mm2. The contributions of our work offer new avenues for inferring force from muscle volume in the context of musculo-skeletal disorder management.
Keywords: Deep convolutional encoder-decoders; Healthy versus pathological transferability; Musculo-skeletal disorders; Obstetrical brachial plexus palsy; Shoulder muscle segmentation.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflicts of interest
The authors declare no conflicts of interest.
Figures
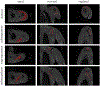
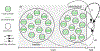
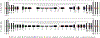

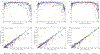




References
-
- Ahmad E, Yap MH, Degens H, McPhee JS, 2014. Atlas-registration based image segmentation of MRI human thigh muscles in 3D space. Medical Imaging: Image Perception Observer Performance, and Technology Assessment.
-
- Andrews S, Hamarneh G, 2015. The generalized log-ratio transformation: learning shape and adjacency priors for simultaneous thigh muscle segmentation. IEEE Trans. Med. Imaging 34 (9), 1773–1787. - PubMed
-
- Barnouin Y, Butler-Browne G, Voit T, Reversat D, Azzabou N, Leroux G, Behin A, McPhee JS, Carlier PG, Hogrel J-Y, 2014. Manual segmentation of individual muscles of the quadriceps femoris using MRI: a reappraisal. J. Magn. Reson. Imaging 40 (1), 239–247. - PubMed
-
- Barra V, Boire J-Y, 2002. Segmentation of fat and muscle from MR images of the thigh by a possibilistic clustering algorithm. Comput. Methods Prog. Biomed 68 (3), 185–193. - PubMed
-
- Baudin P-Y, Azzabou N, Carlier PG, Paragios N, 2012. Prior knowledge, random walks and human skeletal muscle segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention, 569–576. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

